1UYJ
 
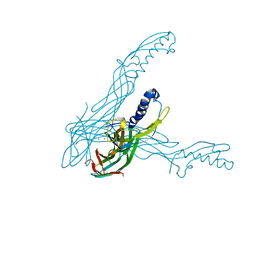 | | Clostridium perfringens epsilon toxin shows structural similarity with the pore forming toxin aerolysin | | Descriptor: | EPSILON-TOXIN, URANIUM ATOM | | Authors: | Cole, A.R, Gibert, M, Poppoff, M, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clostridium Perfringens Epsilon-Toxin Shows Structural Similarity to the Pore-Forming Toxin Aerolysin
Nat.Struct.Mol.Biol., 11, 2004
|
|
3K1L
 
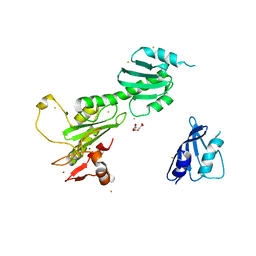 | | Crystal Structure of FANCL | | Descriptor: | CITRIC ACID, Fancl, GOLD ION, ... | | Authors: | Cole, A.R, Walden, H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the catalytic subunit FANCL of the Fanconi anemia core complex
Nat.Struct.Mol.Biol., 17, 2010
|
|
4L5N
 
 | | Crystallographic Structure of HHV-1 Uracil-DNA Glycosylase complexed with the Bacillus phage PZA inhibitor protein p56 | | Descriptor: | ACETATE ION, Early protein GP1B, Uracil-DNA glycosylase | | Authors: | Cole, A.R, Sapir, O, Ryzhenkova, K, Baltulionis, G, Hornyak, P, Savva, R. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Architecturally diverse proteins converge on an analogous mechanism to inactivate Uracil-DNA glycosylase.
Nucleic Acids Res., 41, 2013
|
|
4U0U
 
 | | Wild type eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4U04
 
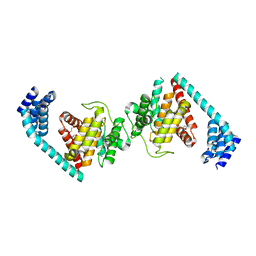 | | Structure of a eukaryotic fic domain containing protein | | Descriptor: | Adenosine monophosphate-protein transferase FICD, D(-)-TARTARIC ACID, TETRAETHYLENE GLYCOL | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4U07
 
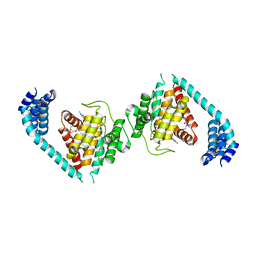 | | ATP bound to eukaryotic FIC domain containing protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4U0Z
 
 | | Eukaryotic Fic Domain containing protein with bound APCPP | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4U0S
 
 | | Structure of Eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Katan, M, Bunney, T.D. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4WJT
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLK
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with reaction product | | Descriptor: | N-[(1R,2S,3R,4R,5R)-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-4-yl]ethanamide, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLI
 
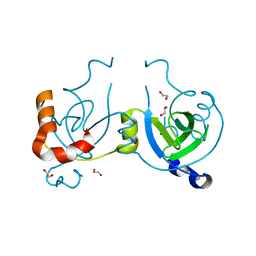 | | Stationary Phase Survival Protein YuiC from B.subtilis | | Descriptor: | 1,2-ETHANEDIOL, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
6YJ2
 
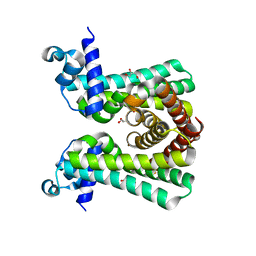 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
6YL2
 
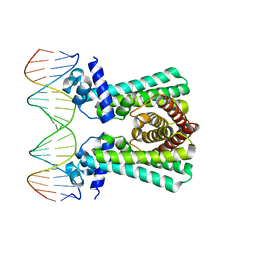 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*TP*AP*AP*TP*GP*AP*CP*GP*AP*TP*TP*AP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*TP*AP*AP*TP*CP*GP*TP*CP*AP*TP*TP*AP*AP*CP*GP*T)-3'), Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
3ZJX
 
 | | Clostridium perfringens epsilon toxin mutant H149A bound to octyl glucoside | | Descriptor: | EPSILON-TOXIN, PHOSPHATE ION, octyl beta-D-glucopyranoside | | Authors: | Bokori-Brown, M, Kokkinidou, M.C, Savva, C.G, Fernandes da Costa, S.P, Naylor, C.E, Cole, A.R, Basak, A.K, Titball, R.W. | | Deposit date: | 2013-01-20 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Clostridium Perfringens Epsilon Toxin H149A Mutant as a Platform for Receptor Binding Studies.
Protein Sci., 22, 2013
|
|
3ZQS
 
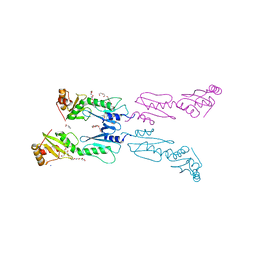 | | Human FANCL central domain | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE FANCL, HEXAETHYLENE GLYCOL, PROLINE, ... | | Authors: | Hodson, C, Cole, A.R, Purkiss-Trew, A, Walden, H. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Fancl, the E3 Ligase in the Fanconi Anemia Pathway.
J.Biol.Chem., 286, 2011
|
|
4H56
 
 | | Crystal structure of the Clostridium perfringens NetB toxin in the membrane inserted form | | Descriptor: | Necrotic enteritis toxin B | | Authors: | Savva, C.G, Fernandes da Costa, S.P, Bokori-Brown, M, Naylor, C, Cole, A.R, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture and Functional Analysis of NetB, a Pore-forming Toxin from Clostridium perfringens.
J.Biol.Chem., 288, 2013
|
|
4KYP
 
 | | Beta-Scorpion Toxin folded in the periplasm of E.coli | | Descriptor: | Beta-insect excitatory toxin Bj-xtrIT, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | O'Reilly, A.O, Cole, A.R, Lopes, J.L, Lampert, A, Wallace, B.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-02-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chaperone-mediated native folding of a beta-scorpion toxin in the periplasm of Escherichia coli.
Biochim.Biophys.Acta, 1840, 2014
|
|
6F2R
 
 | | A heterotetramer of human HspB2 and HspB3 | | Descriptor: | Heat shock protein beta-2, Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-2, HspB2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2, ... | | Authors: | Clark, A.R, Cole, A.R, Boelens, W.C, Keep, N.H, Slingsby, C. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Terminal Regions Confer Plasticity to the Tetrameric Assembly of Human HspB2 and HspB3.
J.Mol.Biol., 430, 2018
|
|
2W2K
 
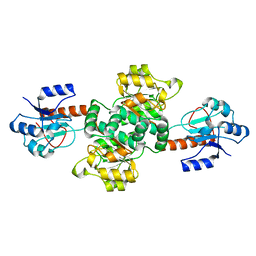 | | Crystal structure of the apo forms of Rhodotorula graminis D- mandelate dehydrogenase at 1.8A. | | Descriptor: | D-MANDELATE DEHYDROGENASE | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
2W2L
 
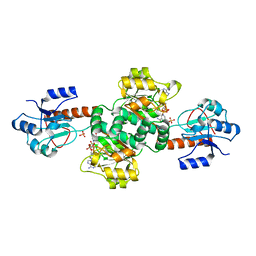 | | Crystal structure of the holo forms of Rhodotorula graminis D- mandelate dehydrogenase at 2.5A. | | Descriptor: | D-MANDELATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
4CAI
 
 | | Structure of inner DysF domain of human dysferlin | | Descriptor: | DYSFERLIN, PHOSPHATE ION | | Authors: | Sula, A, Cole, A.R, Yeats, C, Orengo, C, Keep, N.H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Human Dysferlin Inner Dysf Domain
Bmc Struct.Biol., 14, 2014
|
|
4CAH
 
 | | Structure of inner DysF domain of human dysferlin | | Descriptor: | DYSFERLIN, PHOSPHATE ION | | Authors: | Sula, A, Cole, A.R, Yeats, C, Orengo, C, Keep, N.H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structures of the Human Dysferlin Inner Dysf Domain
Bmc Struct.Biol., 14, 2014
|
|
4F4L
 
 | | Open Channel Conformation of a Voltage Gated Sodium Channel | | Descriptor: | Ion transport protein | | Authors: | McCusker, E.C, Bagneris, C, Naylor, C.E, Cole, A.R, D'Avanzo, N, Nichols, C.G, Wallace, B.A. | | Deposit date: | 2012-05-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing.
Nat Commun, 3, 2012
|
|
4EY0
 
 | | Structure of tandem SH2 domains from PLCgamma1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
4FBN
 
 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
