6GFA
 
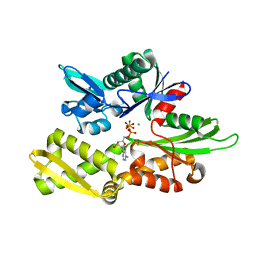 | | Structure of Nucleotide binding domain of HSP110, ATP and Mg2+ complexed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 105 kDa, MAGNESIUM ION | | Authors: | Gonzalez, D, Gotthard, G, Gozzi, G.J, Seigneuric, R, Neiers, F, Briand, L, Garrido, C. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy.
Cell Death Differ., 27, 2020
|
|
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | Descriptor: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | Authors: | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
5OLL
 
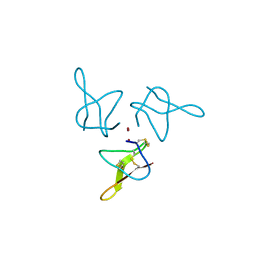 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
8AEH
 
 | |
8AEI
 
 | |
8AEJ
 
 | |
5F0G
 
 | |
3BFH
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with hexadecanoic acid | | Descriptor: | CHLORIDE ION, PALMITIC ACID, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BFB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
2H8V
 
 | | Structure of empty Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Briand, L, Pernollet, J.-C, Cambillau, C, Tegoni, M. | | Deposit date: | 2006-06-08 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational Changes of the Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L upon Ligand Binding
To be Published
|
|
6QQ4
 
 | | Odorant-binding protein dmelOBP28a from Drosophila melanogaster | | Descriptor: | General odorant-binding protein 28a, MALONIC ACID, PENTAETHYLENE GLYCOL | | Authors: | Gonzalez, D, Neiers, F, Gotthard, G, Briand, L. | | Deposit date: | 2019-02-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The Drosophila odorant-binding protein 28a is involved in the detection of the floral odour ss-ionone.
Cell.Mol.Life Sci., 77, 2020
|
|
3BJH
 
 | | Soft-SAD crystal structure of a pheromone binding protein from the honeybee Apis mellifera L. | | Descriptor: | GLYCEROL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Lartigue, A, Gruez, A, Briand, L, Blon, F, Bezirard, V, Walsh, M, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L.
J.Biol.Chem., 279, 2004
|
|
3D75
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D77
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 4.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D74
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, soaked at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D76
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D78
 
 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D73
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3FE6
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand at pH 5.5 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3FE9
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 7.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3FE8
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 4.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3BFA
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CDN
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 4.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CZ1
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
