5IZ2
 
 | | Crystal structure of the N. clavipes spidroin NTD at pH 6.5 | | Descriptor: | Major ampullate spidroin 1A, Major ampullate spidroin 1A (Partial C-terminus) | | Authors: | Atkison, J.H, Olsen, S.K. | | Deposit date: | 2016-03-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
J.Biol.Chem., 291, 2016
|
|
6NYA
 
 | | Crystal Structure of ubiquitin E1 (Uba1) in complex with Ubc3 (Cdc34) and ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6NYO
 
 | | Crystal structure of a human Cdc34-ubiquitin thioester mimetic | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, PHOSPHATE ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6NYD
 
 | |
5UM6
 
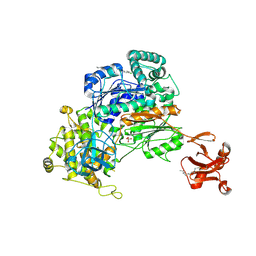 | | Crystal Structure of S. pombe Uba1 in a closed conformation | | Descriptor: | 2-(decylamino)ethane-1-thiol, N-(2-{[(4-chlorophenyl)methyl]disulfanyl}ethyl)decan-1-amine, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Aldana-Masangkay, G, Atkison, J.H, Chen, Y, Olsen, S.K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Domain alternation and active site remodeling are conserved structural features of ubiquitin E1.
J. Biol. Chem., 292, 2017
|
|
6CWY
 
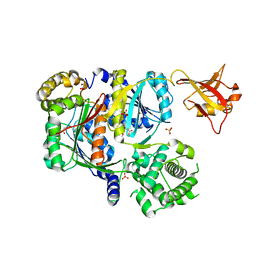 | | Crystal structure of SUMO E1 in complex with an allosteric inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
6CWZ
 
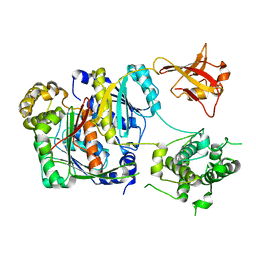 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
6DC6
 
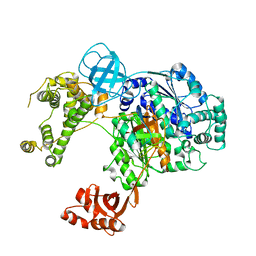 | | Crystal structure of human ubiquitin activating enzyme E1 (Uba1) in complex with ubiquitin | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Ubiquitin, ... | | Authors: | Lv, Z, Yuan, L, Williams, K.M, Atkison, J.H, Olsen, S.K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity.
J. Biol. Chem., 293, 2018
|
|
5TTE
 
 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
