5IHW
 
 | | The crystal structure of SdrE from staphylococcus aureus | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
4QTJ
 
 | | Complex of WOPR domain of Wor1 in Candida albicans with the 13bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
4QTK
 
 | | Complex of WOPR domain of Wor1 in Candida albicans with the 17bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*TP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*AP*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
5G05
 
 | | Cryo-EM structure of combined apo phosphorylated APC | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5G04
 
 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
6Q6H
 
 | |
7N6W
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5H37
 
 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C10 IgG heavy chain variable region, C10 IgG light chain variable region, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lim, X.-N, Ooi, J.S.G, Lambert, S, Tan, T.Y, Widman, D, Shi, J, Baric, R.S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
5H32
 
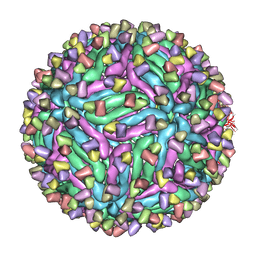 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 5.0 | | Descriptor: | C10 IgG heavy chain variable region, C10 IgG light chain variable region, structural protein E | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
5H30
 
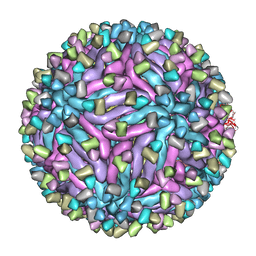 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 6.5 | | Descriptor: | IgG C10 heavy chain, IgG C10 light chain, structural protein E, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
4H5S
 
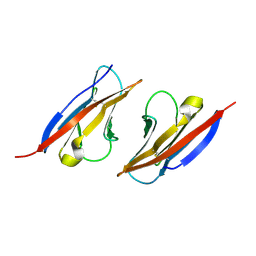 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
8E9X
 
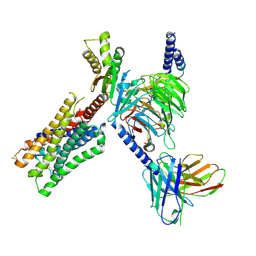 | | CryoEM structure of miniGo-coupled hM4Di in complex with DCZ | | Descriptor: | 11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for selective activation of DREADD-based chemogenetics.
Nature, 612, 2022
|
|
8E9Y
 
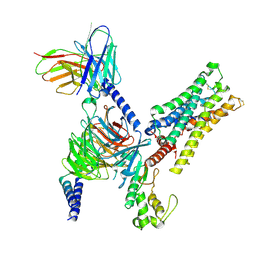 | | CryoEM structure of miniGq-coupled hM3Dq in complex with CNO | | Descriptor: | 8-chloro-11-(4-methyl-4-oxo-4lambda~5~-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis for selective activation of DREADD-based chemogenetics.
Nature, 612, 2022
|
|
8E9W
 
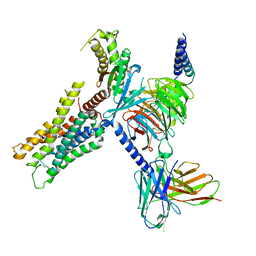 | | CryoEM structure of miniGq-coupled hM3Dq in complex with DCZ | | Descriptor: | 11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis for selective activation of DREADD-based chemogenetics.
Nature, 612, 2022
|
|
8E9Z
 
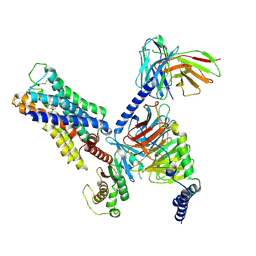 | | CryoEM structure of miniGq-coupled hM3R in complex with Iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis for selective activation of DREADD-based chemogenetics.
Nature, 612, 2022
|
|
8EA0
 
 | | CryoEM structure of miniGq-coupled hM3R in complex with iperoxo (local refinement) | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, CHOLESTEROL HEMISUCCINATE, Muscarinic acetylcholine receptor M3 | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular basis for selective activation of DREADD-based chemogenetics.
Nature, 612, 2022
|
|
3RL1
 
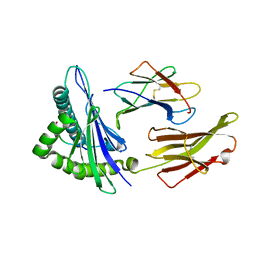 | | HIV RT derived peptide complexed to HLA-A*0301 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-3 alpha chain, ... | | Authors: | Zhang, S, Liu, J, Cheng, H, Tan, S, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2011-04-19 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of cross-allele presentation by HLA-A*0301 and HLA-A*1101 revealed by two HIV-derived peptide complexes
Mol.Immunol., 49, 2011
|
|
3RL2
 
 | | HIV Nef derived peptide Nef73 complexed to HLA-A*0301 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-3 alpha chain, ... | | Authors: | Zhang, S, Liu, J, Cheng, H, Tan, S, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2011-04-19 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Structural basis of cross-allele presentation by HLA-A*0301 and HLA-A*1101 revealed by two HIV-derived peptide complexes
Mol.Immunol., 49, 2011
|
|
6W7H
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 6-methyl-2-oxo-2,5-dihydropyrimidine-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Augelli-Szafran, C.E, Pathak, A.K, Wu, M. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W8K
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 5,6-dimethyl-2-oxo-2,3-dihydropyrimidine-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Augelli-Szafran, C.E, Pathak, A.K, Wu, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
