1HG3
 
 | | Crystal structure of tetrameric TIM from Pyrococcus woesei. | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Bell, G.S, Russell, R.J.M, Siebers, B, Hensel, R, Taylor, G.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tiny Tim: A Small, Tetrameric, Hyperthermostable Triosephosphate Isomerase
J.Mol.Biol., 306, 2001
|
|
1KY8
 
 | | Crystal Structure of the Non-phosphorylating glyceraldehyde-3-phosphate Dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pohl, E, Brunner, N, Wilmanns, M, Hensel, R. | | Deposit date: | 2002-02-04 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Allosteric Non-phosphorylating
glyceraldehyde-3-phosphate Dehydrogenase from the Hyperthermophilic
Archaeum Thermoproteus tenax
J.Biol.Chem., 277, 2002
|
|
1LLC
 
 | | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS CASEI AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, L-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Buehner, M, Hecht, H.J, Hensel, R. | | Deposit date: | 1988-11-21 | | Release date: | 1989-07-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS-CASEI AT 3A RESOLUTION.
Acta Crystallogr.,Sect.A, 40, 1984
|
|
2YCE
 
 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
4AY7
 
 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AY8
 
 | | SeMet-derivative of a methyltransferase from M. mazei | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-THIOETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeldt, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1OJX
 
 | | Crystal structure of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-16 | | Release date: | 2003-09-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK6
 
 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK4
 
 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1UXQ
 
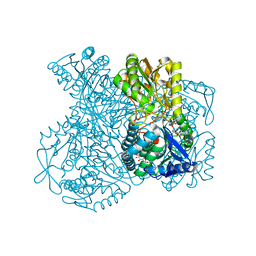 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXP
 
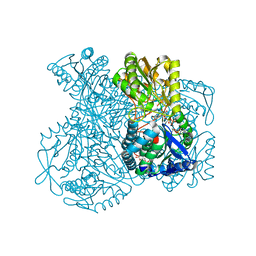 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXN
 
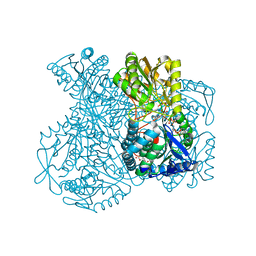 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXR
 
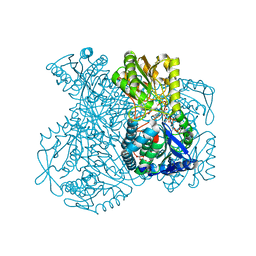 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXT
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXU
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1W0M
 
 | | Triosephosphate isomerase from Thermoproteus tenax | | Descriptor: | PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Taylor, G, Lorentzen, E, Pohl, E, Lilie, H, Schramm, A, Knura, T, Stubbe, K, Tjaden, B, Hensel, R. | | Deposit date: | 2004-06-08 | | Release date: | 2004-09-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of a Regulated Archaeal Triosephosphate Isomerase Adapted to High Temperature
J.Mol.Biol., 342, 2004
|
|
1W8S
 
 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
