2BI8
 
 | |
2BI7
 
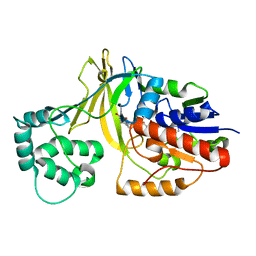 | | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Srikannathasan, V, Naismith, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mycobacteria Tuberculosis and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the Oxidised State and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the (Active) Reduced State.
J.Mol.Biol., 348, 2005
|
|
1V0J
 
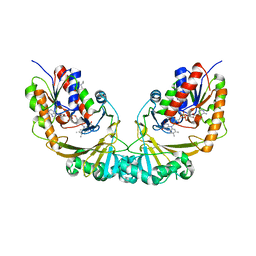 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
1WAM
 
 | |
1OC2
 
 | |
7Z6F
 
 | |
5OFR
 
 | |
5OFP
 
 | |
6STL
 
 | | Taurine ABC transporter substrate binding protein TauA from E. coli in complex with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine-binding periplasmic protein | | Authors: | Beis, K, Qu, F, Wagner, A, ElOmari, K. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Desolvation of the substrate-binding protein TauA dictates ligand specificity for the alkanesulfonate ABC importer TauABC.
Biochem.J., 476, 2019
|
|
6SSY
 
 | |
6ST1
 
 | |
6ST0
 
 | |
7ZVS
 
 | |
6RCK
 
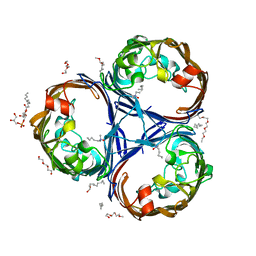 | | Crystal structure of the OmpK36 GD insertion chimera from Klebsiella pneumonia | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Beis, K, Romano, M, Kwong, J. | | Deposit date: | 2019-04-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo.
Nat Commun, 10, 2019
|
|
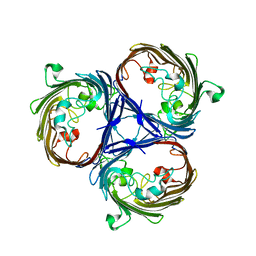
6RCP
 
 | |
6RD3
 
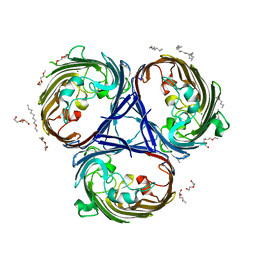 | |
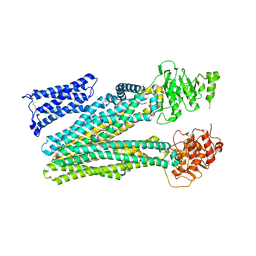
8RQ4
 
 | |
6YO1
 
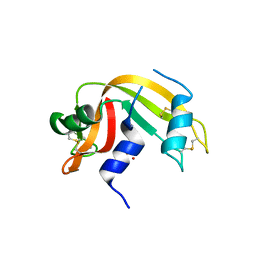 | | Crystal structure of ribonuclease A solved by vanadium SAD phasing | | Descriptor: | Ribonuclease pancreatic, URIDINE-2',3'-VANADATE | | Authors: | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
6YSO
 
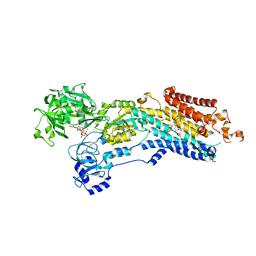 | | Crystal structure of the (SR) Ca2+-ATPase solved by vanadium SAD phasing | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-11-04 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
5EG1
 
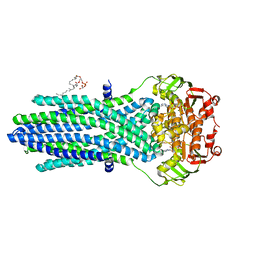 | | Antibacterial peptide ABC transporter McjD with a resolved lipid | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, ... | | Authors: | Choudhury, H.G, Beis, K. | | Deposit date: | 2015-10-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural and Functional Basis for Lipid Synergy on the Activity of the Antibacterial Peptide ABC Transporter McjD.
J.Biol.Chem., 291, 2016
|
|
4PL0
 
 | |
4YCR
 
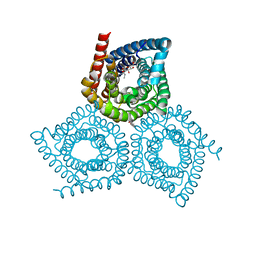 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | Descriptor: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | Authors: | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7P34
 
 | |
7PZF
 
 | |
7Q3T
 
 | |
