1DVW
 
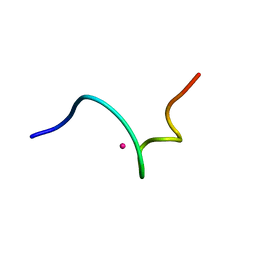 | | NMR structure of 18 residue peptide from merp protein | | Descriptor: | 18 RESIDUE PEPTIDE FROM MERP PROTEIN, MERCURY (II) ION | | Authors: | Veglia, G, Porcelli, F, De Silva, T.M, Prantner, A.M, Opella, S.J. | | Deposit date: | 2000-01-22 | | Release date: | 2003-12-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Metal-Binding Motif
GMTCAAC Is Similar in an 18-Residue Linear
Peptide and the Mercury Binding Protein MerP
J.Am.Chem.Soc., 122, 2000
|
|
1JDM
 
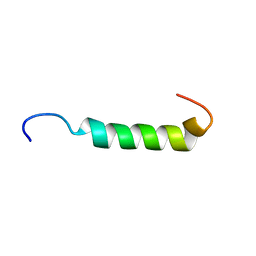 | | NMR Structure of Sarcolipin | | Descriptor: | Sarcolipin | | Authors: | Veglia, G, Mascioni, A. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of sarcolipin in lipid environments.
Biochemistry, 41, 2002
|
|
1ZZA
 
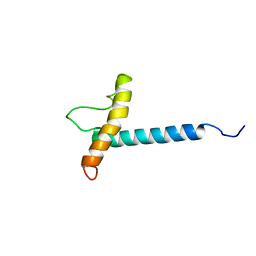 | | Solution NMR Structure of the Membrane Protein Stannin | | Descriptor: | Stannin | | Authors: | Buck-Koehntop, B.A, Mascioni, A, Buffy, J.J, Veglia, G. | | Deposit date: | 2005-06-13 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride.
J.Mol.Biol., 354, 2005
|
|
1XC0
 
 | | Twenty Lowest Energy Structures of Pa4 by Solution NMR | | Descriptor: | Pardaxin P-4 | | Authors: | Porcelli, F, Buck, B, Lee, D.-K, Hallock, K.J, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of pardaxin determined by NMR experiments in model membranes
J.Biol.Chem., 279, 2004
|
|
1N7L
 
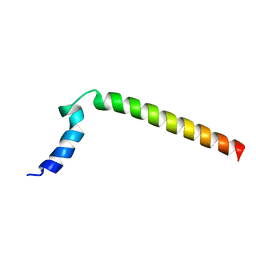 | |
2KB7
 
 | | Hybrid solution and solid-state NMR structure of monomeric phospholamban in lipid bilayers | | Descriptor: | Phospholamban | | Authors: | Traaseth, N.J, Shi, L, Verardi, R, Veglia, G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2021-10-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
1KVI
 
 | |
1KVJ
 
 | |
1KUW
 
 | | High-Resolution Structure and Localization of Amylin Nucleation Site in Detergent Micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Mascioni, A, Porcelli, F, Ilangovan, U, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2002-01-22 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences of the amylin nucleation site in SDS micelles: an NMR study.
Biopolymers, 69, 2003
|
|
2KYV
 
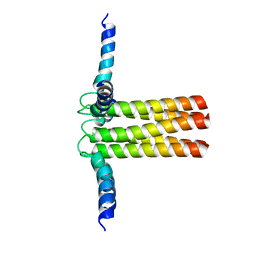 | | Hybrid solution and solid-state NMR structure ensemble of phospholamban pentamer | | Descriptor: | Phospholamban | | Authors: | Verardi, R, Shi, L, Traaseth, N.J, Veglia, G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LPF
 
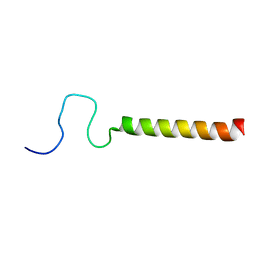 | | R state structure of monomeric phospholamban (C36A, C41F, C46A) | | Descriptor: | Cardiac phospholamban | | Authors: | De Simone, A, Montalvao, R.W, Gustavsson, M, Shi, L, Veglia, G, Vendruscolo, M. | | Deposit date: | 2012-02-11 | | Release date: | 2013-02-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of the Excited States of Phospholamban and Shifts in Their Populations upon Phosphorylation.
Biochemistry, 52, 2013
|
|
2LZE
 
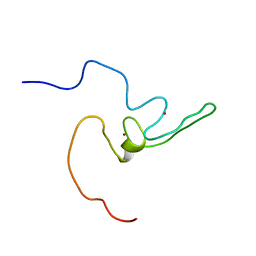 | | Ligase 10C | | Descriptor: | ZINC ION, a primordial catalytic fold generated by in vitro evolution | | Authors: | Chao, F, Morelli, A, Haugner, J, Churchfield, L, Hagmann, L, Shi, L, Masterson, L, Sarangi, R, Veglia, G, Seelig, B. | | Deposit date: | 2012-10-01 | | Release date: | 2012-10-24 | | Last modified: | 2013-02-06 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of a primordial catalytic fold generated by in vitro evolution.
Nat.Chem.Biol., 9, 2013
|
|
2M3B
 
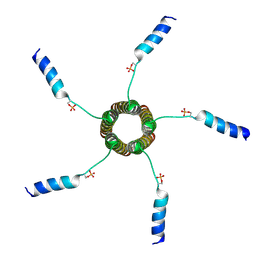 | |
2MZ0
 
 | |
7MPA
 
 | |
3O7L
 
 | |
6E21
 
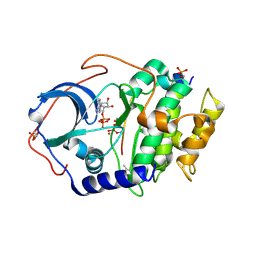 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
2XJH
 
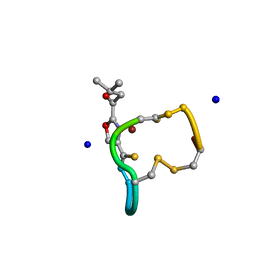 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2XJI
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
