2VS0
 
 | |
2VRZ
 
 | |
6FTX
 
 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2018-10-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
6G0L
 
 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
2IYP
 
 | | product rup | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, ADENOSINE-2'-5'-DIPHOSPHATE, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of a Bacterial 6-Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
2IZ1
 
 | | 6PDH complexed with PEX inhibitor synchrotron data | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-23 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Bacterial 6- Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
2IZ0
 
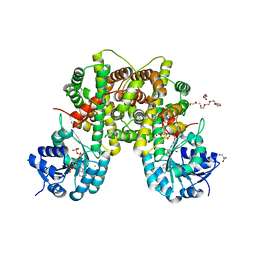 | | PEX inhibitor-home data | | Descriptor: | 1,2-ETHANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-23 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of a Bacterial 6- Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
2IYO
 
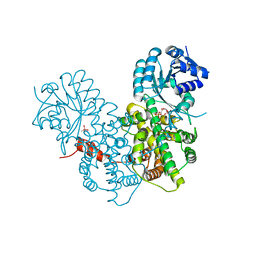 | | Structural characterization of a bacterial 6PDH reveals aspects of specificity, mechanism and mode of inhibition | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, 6-PHOSPHOGLUCONIC ACID, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of a Bacterial 6-Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
2C7Z
 
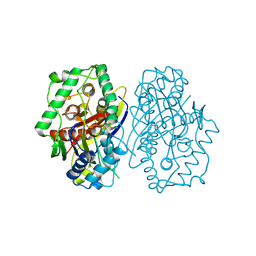 | | Plant enzyme crystal form II | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
2C7Y
 
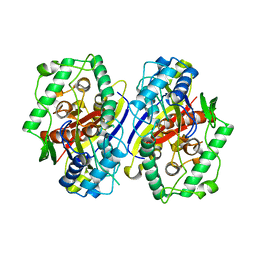 | | plant enzyme | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Germain, V, Bryce, J.H, Smith, S.M, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
8OV6
 
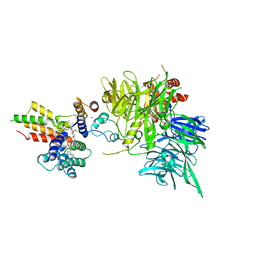 | | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DDB1deltaBPB, ... | | Authors: | Cowan, A.D, Sundaramoorthy, R, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Targeted protein degradation via intramolecular bivalent glues.
Nature, 627, 2024
|
|
1FBZ
 
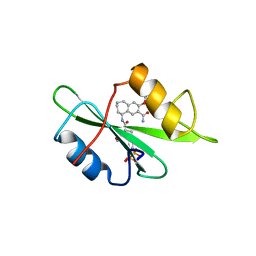 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
5N2W
 
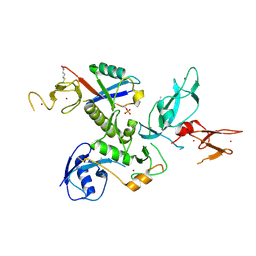 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
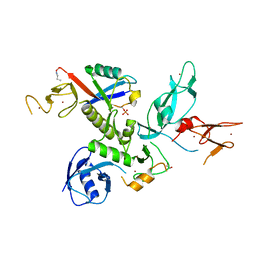 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8B2X
 
 | |
8ANE
 
 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
2XB0
 
 | |
5C23
 
 | | Parkin (S65DUblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5C1Z
 
 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5AGC
 
 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
1IJR
 
 | | Crystal structure of LCK SH2 complexed with nonpeptide phosphotyrosine mimetic | | Descriptor: | (4-{2-ACETYLAMINO-2-[1-(3-CARBAMOYL-4-CYCLOHEXYLMETHOXY-PHENYL)-ETHYLCARBAMOYL}-ETHYL}-2-PHOSPHONO-PHENOXY)-ACETIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Kawahata, N.H, Yang, M.H, Luke, G.P, Shakespeare, W.C, Sundaramoorthi, R. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel phosphotyrosine mimetic 4'-carboxymethyloxy-3'-phosphonophenylalanine (Cpp): exploitation in the design of nonpeptide inhibitors of pp60(Src) SH2 domain.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
5O6C
 
 | | Crystal Structure of a threonine-selective RCR E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2, ZINC ION | | Authors: | Pao, K.-C, Rafie, K.Z, van Aalten, D, Virdee, S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Nature, 556, 2018
|
|
3IK3
 
 | |
2BDJ
 
 | | Src kinase in complex with inhibitor AP23464 | | Descriptor: | 3-[2-(2-CYCLOPENTYL-6-{[4-(DIMETHYLPHOSPHORYL)PHENYL]AMINO}-9H-PURIN-9-YL)ETHYL]PHENOL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dalgarno, D, Stehle, T, Schelling, P, Narula, S, Sawyer, T. | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.
Chem.Biol.Drug Des., 67, 2006
|
|
