2CJJ
 
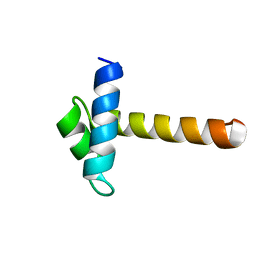 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
3ZPL
 
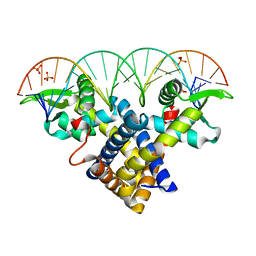 | | Crystal structure of Sco3205, a MarR family transcriptional regulator from Streptomyces coelicolor, in complex with DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*TP*GP*AP*GP*AP*TP*CP*TP *CP*AP*AP*TP*CP*TP*TP*DT)-3', PHOSPHATE ION, PUTATIVE MARR-FAMILY TRANSCRIPTIONAL REPRESSOR | | Authors: | Stevenson, C.E.M, Assaad, A, Lawson, D.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of DNA Sequence Recognition by a Streptomycete Marr Family Transcriptional Regulator Through Surface Plasmon Resonance and X-Ray Crystallography.
Nucleic Acids Res., 41, 2013
|
|
1E5K
 
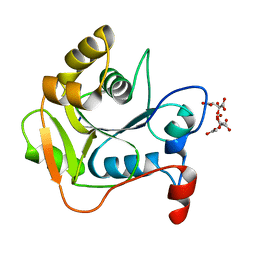 | | CRYSTAL STRUCTURE OF THE MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN MOBA (PROTEIN FA) FROM ESCHERICHIA COLI AT NEAR ATOMIC RESOLUTION | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Stevenson, C.E.M, Sargent, F, Buchanan, G, Palmer, T, Lawson, D.M. | | Deposit date: | 2000-07-27 | | Release date: | 2000-11-07 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Molybdenum Cofactor Biosynthesis Protein Moba from Escherichia Coli at Near Atomic Resolution
Structure, 8, 2000
|
|
3ZMD
 
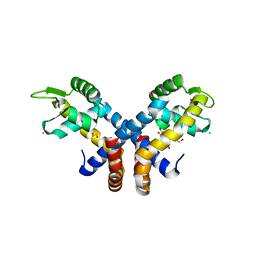 | | Crystal structure of AbsC, a MarR family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stevenson, C.E.M, Kock, H, Mootien, S, Davies, S.C, Bibb, M.J, Lawson, D.M. | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Absc, a Marr Family Transcriptional Regulator from Streptomyces Coelicolor
To be Published
|
|
7P2X
 
 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | Descriptor: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
4BWV
 
 | | Structure of Adenosine 5-prime-phosphosulfate Reductase apr-b from Physcomitrella Patens | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHOADENOSINE-PHOSPHOSULPHATE REDUCTASE | | Authors: | Stevenson, C.E.M, Hughes, R.K, McManus, M.T, Lawson, D.M, Kopriva, S. | | Deposit date: | 2013-07-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Crystal Structure of Apr-B, an Atypical Adenosine 5-Prime-Phosphosulfate Reductase from Physcomitrella Patens
FEBS Lett., 587, 2013
|
|
7P2N
 
 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
7P2M
 
 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
8PFC
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFD
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6HQ8
 
 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 with laminarihexaose bound at a surface site | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
6HQ6
 
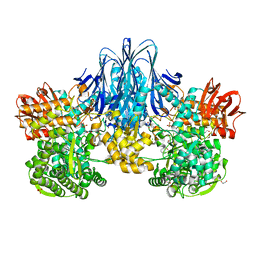 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Bacterial beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
4WUB
 
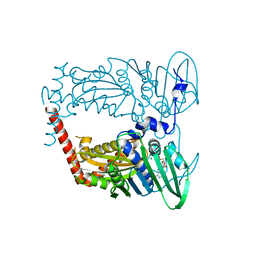 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WUC
 
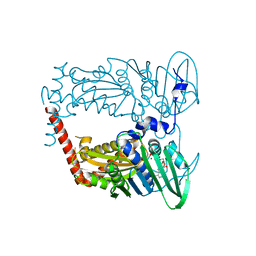 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2UYJ
 
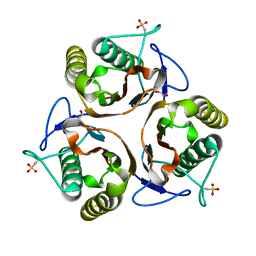 | | Crystal structure of E. coli TdcF with bound ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYA
 
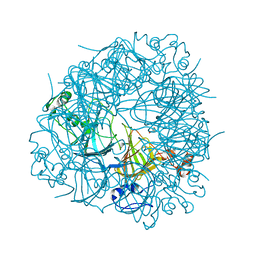 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
8BV0
 
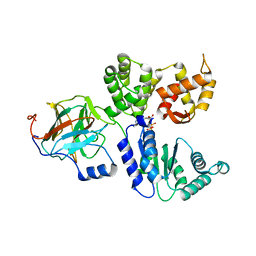 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
8BN6
 
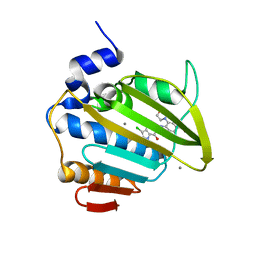 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
3ZT7
 
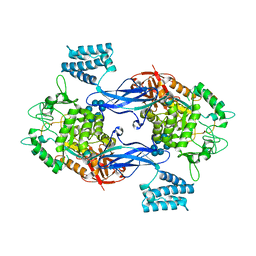 | | GlgE isoform 1 from Streptomyces coelicolor with beta-cyclodextrin and maltose bound | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT5
 
 | | GlgE isoform 1 from Streptomyces coelicolor with maltose bound | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT6
 
 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin and maltose bound | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
4WUD
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
