8EOB
 
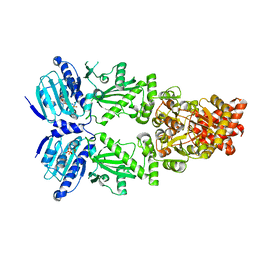 | | Cryo-EM structure of human HSP90B in the closed state | | Descriptor: | Heat shock protein HSP 90-beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
8EOA
 
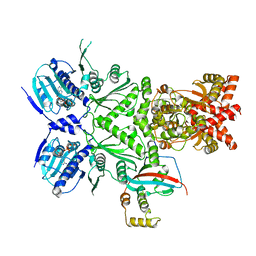 | | Cryo-EM structure of human HSP90B-AIPL1 complex | | Descriptor: | Aryl-hydrocarbon-interacting protein-like 1, Heat shock protein HSP 90-beta, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
6N84
 
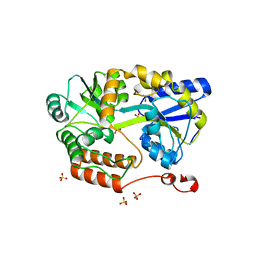 | | MBP-fusion protein of transducin-alpha residues 327-350 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Srivastava, D, Gakhar, L, Artemyev, N.O. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
6N86
 
 | |
6NUB
 
 | | Pyruvate Kinase M2 Mutant - S437Y in Complex with L-serine | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
6NU1
 
 | | Crystal Structure of Human PKM2 in Complex with L-cysteine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CYSTEINE, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
6NU5
 
 | | Pyruvate Kinase M2 Mutant - S437Y in Complex with L-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
6N85
 
 | | Resistance to inhibitors of cholinesterase 8A (Ric8A) protein in complex with MBP-tagged transducin-alpha residues 327-350 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, Synembryn-A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Srivastava, D, Gakhar, L, Artemyev, N.O. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-10 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
6B6U
 
 | | Pyruvate Kinase M2 mutant - S437Y | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Srivastava, D, Dey, M. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of a Dimeric Variant of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 56, 2017
|
|
7UMO
 
 | | Structure of Unc119-inhibitor complex. | | Descriptor: | (3s,5s,7s)-N-(4,5-dichloropyridin-2-yl)adamantane-1-carboxamide, GLYCEROL, Protein unc-119 homolog A | | Authors: | Srivastava, D, Sebag, J.A, Artemyev, N.O. | | Deposit date: | 2022-04-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin sensitization by small molecules enhancing GLUT4 translocation.
Cell Chem Biol, 30, 2023
|
|
3V9K
 
 | |
3V9L
 
 | |
3V9G
 
 | |
3V9J
 
 | |
6WP3
 
 | | Pyruvate Kinase M2 Mutant-K433Q | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6WP5
 
 | | Pyruvate Kinase M2 mutant-S37D | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6WP6
 
 | | Pyruvate Kinase M2 mutant-S37E K433E | | Descriptor: | GLYCEROL, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6WP4
 
 | | Pyruvate Kinase M2 mutant-S37E | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
3HAZ
 
 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ITG
 
 | |
3V9I
 
 | |
3V9H
 
 | |
8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
4H6Q
 
 | |
4H6R
 
 | | Structure of reduced Deinococcus radiodurans proline dehydrogenase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Proline dehydrogenase | | Authors: | Min, L, Tanner, J.J. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release.
Biochemistry, 51, 2012
|
|
