1GD1
 
 | |
2GD1
 
 | |
1UAE
 
 | | STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Skarzynski, T. | | Deposit date: | 1996-09-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure, 4, 1996
|
|
1A2N
 
 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
1GQQ
 
 | |
1GQY
 
 | | MURC - CRYSTAL STRUCTURE OF THE ENZYME FROM HAEMOPHILUS INFLUENZAE COMPLEXED WITH AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE | | Authors: | Skarzynski, T, Cleasby, A, Domenici, E, Gevi, M, Shaw, J. | | Deposit date: | 2001-12-06 | | Release date: | 2003-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Udp-N-Acetylmuramate-L-Alanine Ligase (Murc) from Haemophilus Influenzae
To be Published
|
|
1DTX
 
 | |
1RTL
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN: NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND PYRROLIDINE-5,5-TRANSLACTAM INHIBITOR | | Descriptor: | N-[(2R,3S)-1-((2S)-2-{[(CYCLOPENTYLAMINO)CARBONYL]AMINO}-3-METHYLBUTANOYL)-2-(1-FORMYL-1-CYCLOBUTYL)PYRROLIDINYL]CYCLOPROPANECARBOXAMIDE, NS3 protease/helicase, NS4A COFACTOR, ... | | Authors: | Skarzynski, T, Somers, D.O.N. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Org.Lett., 5, 2003
|
|
1HV7
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH GW311616A | | Descriptor: | ELASTASE 1, N-[2-(1-FORMYL-2-METHYL-PROPYL)-1-(4-PIPERIDIN-1-YL-BUT-2-ENOYL)-PYRROLIDIN-3-YL]-METHANESULFONAMIDE, SULFATE ION | | Authors: | Skarzynski, T, Singh, O.M. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of a potent, intracellular, orally bioavailable, long duration inhibitor of human neutrophil elastase--GW311616A a development candidate
Bioorg.Med.Chem.Lett., 11, 2001
|
|
2Z2C
 
 | | MURA inhibited by unag-cnicin adduct | | Descriptor: | 2-acetamido-3-O-[(2S,3S)-2-carboxy-3,4-dihydroxybutan-2-yl]-2-deoxy-alpha-D-glucopyranose, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Steinbach, A, Skarzynski, T, Scheidig, A.J, Klein, C.D. | | Deposit date: | 2007-05-17 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Antibacterial Target Enzyme Mura Synthesizes its Own Non-Covalent Suicide Inhibitor from the Natural Product Cnicin
To be Published
|
|
1N1L
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND INHIBITOR (GW472467X) | | Descriptor: | HCV NS3 SERINE PROTEASE, NS4A COFACTOR, ZINC ION, ... | | Authors: | Andrews, D.M, Chaignot, H, Coomber, B.A, Good, A.C, Hind, S.L, Jones, P.S, Mill, G, Robinson, J.E, Skarzynski, T, Slater, M.J, Somers, D.O.N. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray Crystal Structure Data in the Optimisation of P3 and P4 Substituents
Org.Lett., 4, 2002
|
|
1PMI
 
 | | Candida Albicans Phosphomannose Isomerase | | Descriptor: | PHOSPHOMANNOSE ISOMERASE, ZINC ION | | Authors: | Cleasby, A, Skarzynski, T, Wonacott, A, Davies, G.J, Hubbard, R.E, Proudfoot, A.E.I, Wells, T.N.C, Payton, M.A, Bernard, A.R. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
1DUH
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
4BHN
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIC
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIE
 
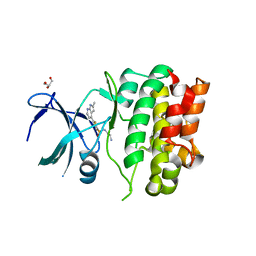 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BF2
 
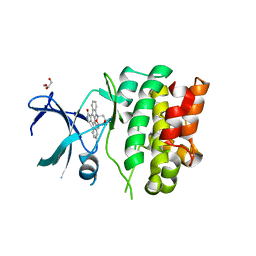 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BID
 
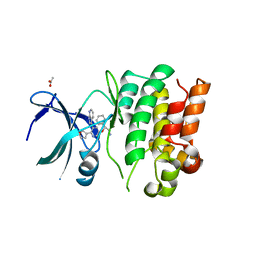 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 5-[(1R)-2-amino-1-phenylethoxy]-2-(furan-3-yl)-3-{1H-pyrrolo[2,3-b]pyridin-3-yl}pyridine, ACETATE ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIB
 
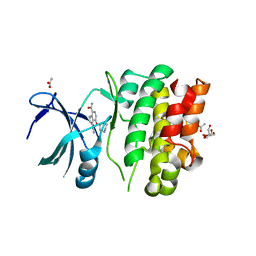 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 3-cyano-4-(piperidin-4-yloxy)-1H-indole-7-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
1A4G
 
 | | INFLUENZA VIRUS B/BEIJING/1/87 NEURAMINIDASE COMPLEXED WITH ZANAMIVIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Cleasby, A, Singh, O, Skarzynski, T, Wonacott, A.J. | | Deposit date: | 1998-01-29 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B.
J.Med.Chem., 41, 1998
|
|
1A4Q
 
 | | INFLUENZA VIRUS B/BEIJING/1/87 NEURAMINIDASE COMPLEXED WITH DIHYDROPYRAN-PHENETHYL-PROPYL-CARBOXAMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ACETYLAMINO-4-AMINO-6-(PHENETHYL-PROPYL-CARBAMOYL)-5,6-DIHYDRO-4H-PYRAN-2-CARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Singh, O, Skarzynski, T, Wonacott, A.J. | | Deposit date: | 1998-01-30 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B.
J.Med.Chem., 41, 1998
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1FBL
 
 | | STRUCTURE OF FULL-LENGTH PORCINE SYNOVIAL COLLAGENASE (MMP1) REVEALS A C-TERMINAL DOMAIN CONTAINING A CALCIUM-LINKED, FOUR-BLADED BETA-PROPELLER | | Descriptor: | CALCIUM ION, FIBROBLAST (INTERSTITIAL) COLLAGENASE (MMP-1), N-[3-(N'-HYDROXYCARBOXAMIDO)-2-(2-METHYLPROPYL)-PROPANOYL]-O-TYROSINE-N-METHYLAMIDE, ... | | Authors: | Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1995-04-24 | | Release date: | 1996-01-29 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller.
Structure, 3, 1995
|
|
1RTG
 
 | |
5TSO
 
 | |
