2F1K
 
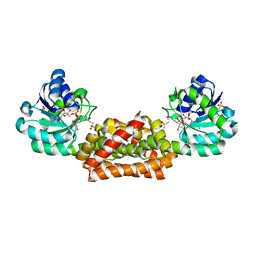 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
5E7F
 
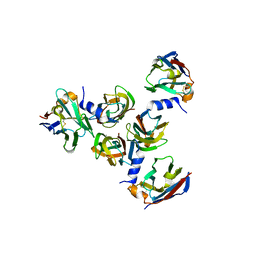 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7T
 
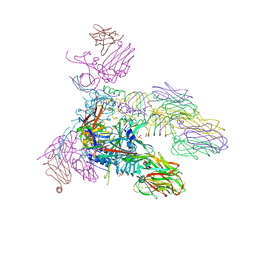 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7B
 
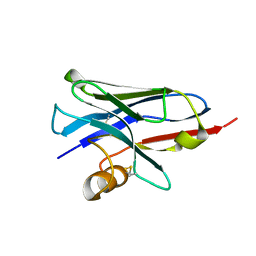 | | Structure of a nanobody (vHH) from camel against phage Tuc2009 RBP (BppL, ORF53) | | Descriptor: | nanobody nano-L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
2UY1
 
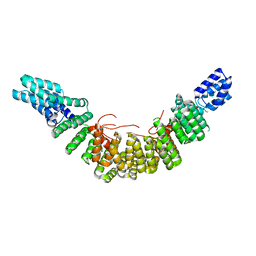 | |
4A6E
 
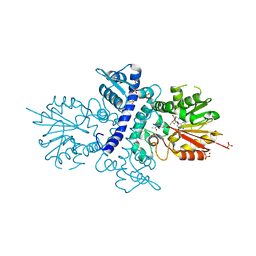 | | Crystal structure of human N-acetylserotonin methyltransferase (ASMT) in complex with SAM and N-acetylserotonin | | Descriptor: | GLYCEROL, HYDROXYINDOLE O-METHYLTRANSFERASE, N-ACETYL SEROTONIN, ... | | Authors: | Legrand, P, Haouz, A, Shepard, W. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Mapping of Human Asmt, the Last Enzyme of the Melatonin Synthesis Pathway.
J.Pineal Res., 54, 2013
|
|
4A6D
 
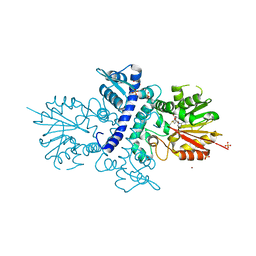 | | Crystal structure of human N-acetylserotonin methyltransferase (ASMT) in complex with SAM | | Descriptor: | GLYCEROL, HYDROXYINDOLE O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE, ... | | Authors: | Legrand, P, Haouz, A, Shepard, W. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Mapping of Human Asmt, the Last Enzyme of the Melatonin Synthesis Pathway.
J.Pineal Res., 54, 2013
|
|
2V94
 
 | |
7QXM
 
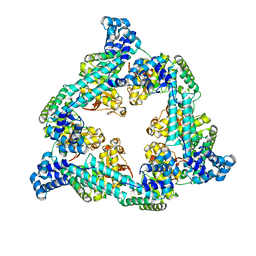 | |
8AAJ
 
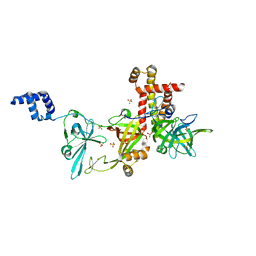 | | Crystal structure of the Pyrococcus abyssi RPA (apo form) | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Legrand, P, Madru, C, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
4D5M
 
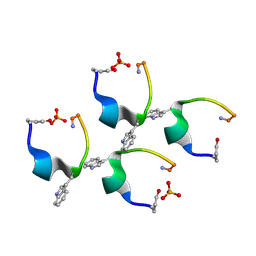 | | Gonadotropin-releasing hormone agonist | | Descriptor: | PHOSPHATE ION, TRIPTORELIN | | Authors: | Legrand, P, Le Du, M.-H, Valery, C, Deville-Foillard, S, Paternostre, M, Artzner, F. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Atomic View of the Histidine Environment Stabilizing Higher- Ph Conformations of Ph-Dependent Proteins.
Nat.Commun., 6, 2015
|
|
6T66
 
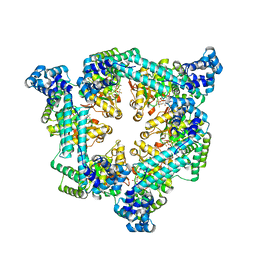 | | Crystal structure of the Vibrio cholerae replicative helicase (DnaB) with GDP-AlF4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Legrand, P, Quevillon-Cheruel, S, Li de la Sierra-Gallay, I, Walbott, H. | | Deposit date: | 2019-10-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res., 49, 2021
|
|
3Q4F
 
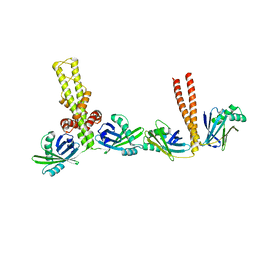 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4UUZ
 
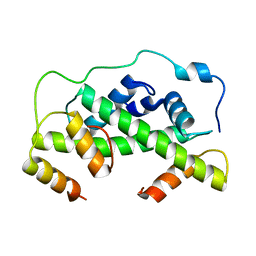 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
1XJB
 
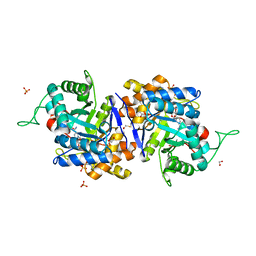 | | Crystal structure of human type 3 3alpha-hydroxysteroid dehydrogenase in complex with NADP(H), citrate and acetate molecules | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C2, ... | | Authors: | Couture, J.-F, Pereira de Jesus-Tran, K, Roy, A.-M, Legrand, P, Cantin, L, Cote, P.-L, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2004-09-23 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of crystal structures of human type 3 3alpha-hydroxysteroid dehydrogenase reveals an "induced-fit" mechanism and a conserved basic motif involved in the binding of androgen
Protein Sci., 14, 2005
|
|
2PNU
 
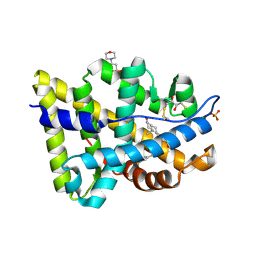 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
3CAO
 
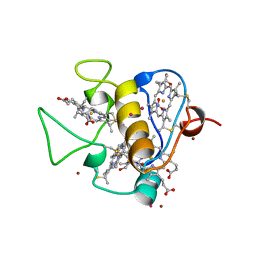 | | OXIDISED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
3CAR
 
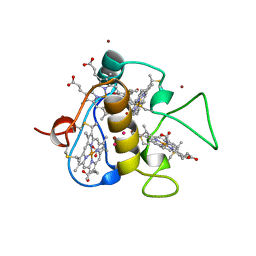 | | REDUCED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
7NXR
 
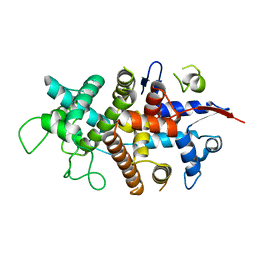 | |
7NXQ
 
 | |
7NXP
 
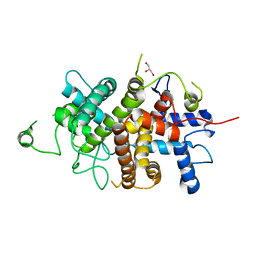 | |
1EYX
 
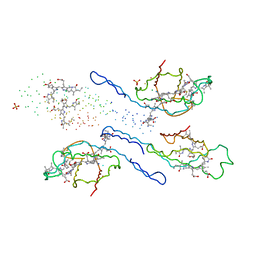 | | CRYSTAL STRUCTURE OF R-PHYCOERYTHRIN AT 2.2 ANGSTROMS | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOUROBILIN, ... | | Authors: | Contreras-Martel, C, Legrand, P, Piras, C, Vernede, X, Martinez-Oyanedel, J, Bunster, M, Fontecilla-Camps, J.C. | | Deposit date: | 2000-05-09 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallization and 2.2 A resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4YO3
 
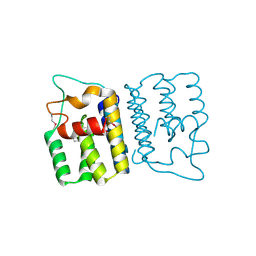 | | Enteroaggregative Escherichia Coli TssA N-terminal fragment | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
4YO5
 
 | | EAEC T6SS TssA-Cterminus | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
