2PJR
 
 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
8B1T
 
 | | RecBCD-DNA in complex with the phage protein Abc2 | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8B1R
 
 | | RecBCD in complex with the phage protein gp5.9 | | Descriptor: | MAGNESIUM ION, Probable RecBCD inhibitor gp5.9, RecBCD enzyme subunit RecB, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8B1U
 
 | | RecBCD-DNA in complex with the phage protein Abc2 and host PpiB | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
3PJR
 
 | | HELICASE SUBSTRATE COMPLEX | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism
Cell(Cambridge,Mass.), 97, 1999
|
|
1W36
 
 | | RecBCD:DNA complex | | Descriptor: | CALCIUM ION, DNA HAIRPIN, EXODEOXYRIBONUCLEASE V ALPHA CHAIN, ... | | Authors: | Singleton, M.R, Dillingham, M.S, Gaudier, M, C Kowalczykowski, S, Wigley, D.B. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Recbcd Enzyme Reveals a Machine for Processing DNA Breaks
Nature, 432, 2004
|
|
1QHH
 
 | | STRUCTURE OF DNA HELICASE WITH ADPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (PCRA (SUBUNIT)) | | Authors: | Soultanas, P, Dillingham, M.S, Velankar, S.S, Wigley, D.B. | | Deposit date: | 1999-05-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase.
J.Mol.Biol., 290, 1999
|
|
1QHG
 
 | | STRUCTURE OF DNA HELICASE MUTANT WITH ADPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HELICASE PCRA, MAGNESIUM ION | | Authors: | Soultanas, P, Dillingham, M.S, Velankar, S.S, Wigley, D.B. | | Deposit date: | 1999-05-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase.
J.Mol.Biol., 290, 1999
|
|
5NOC
 
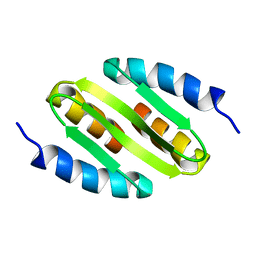 | |
5DMA
 
 | |
5MBV
 
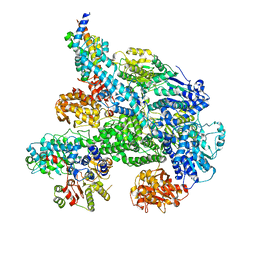 | | Cryo-EM structure of Lambda Phage protein GamS bound to RecBCD. | | Descriptor: | Host-nuclease inhibitor protein gam, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-11-08 | | Release date: | 2017-01-11 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones.
Elife, 5, 2016
|
|
3U44
 
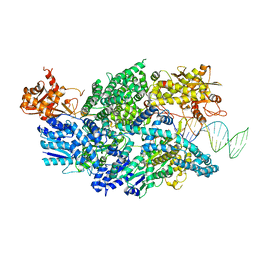 | | Crystal structure of AddAB-DNA complex | | Descriptor: | ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, DNA (36-MER), ... | | Authors: | Saikrishnan, K, Krajewski, W, Wigley, D. | | Deposit date: | 2011-10-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
3U4Q
 
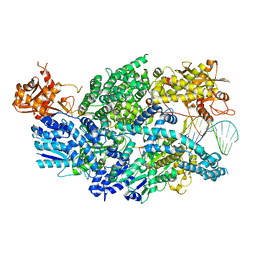 | | Structure of AddAB-DNA complex at 2.8 angstroms | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, ... | | Authors: | Saikrishnan, K, Krajewski, W, Wigley, D. | | Deposit date: | 2011-10-10 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
4CEH
 
 | | Crystal structure of AddAB with a forked DNA substrate | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEI
 
 | | Crystal structure of ADPNP-bound AddAB with a forked DNA substrate | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEJ
 
 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
6G1C
 
 | |
6G1N
 
 | |
6G26
 
 | | The crystal structure of the Burkholderia pseudomallei HicAB complex | | Descriptor: | 1,2-ETHANEDIOL, HicA, HicB, ... | | Authors: | Winter, A.J, Isupov, M.N, Williams, C, Crump, M.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J. Biol. Chem., 293, 2018
|
|
3K70
 
 | |
