2ZNB
 
 | | METALLO-BETA-LACTAMASE (CADMIUM-BOUND FORM) | | Descriptor: | CADMIUM ION, METALLO-BETA-LACTAMASE, SODIUM ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 6, 1997
|
|
3ZNB
 
 | | METALLO-BETA-LACTAMASE (ZN, HG-BOUND FORM) | | Descriptor: | MERCURY (II) ION, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1997-10-15 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 6, 1997
|
|
1DDK
 
 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
2RAN
 
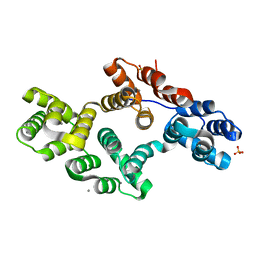 | | RAT ANNEXIN V CRYSTAL STRUCTURE: CA2+-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Concha, N.O, Head, J.F, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1994-09-01 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Rat annexin V crystal structure: Ca(2+)-induced conformational changes.
Science, 261, 1993
|
|
1ZNB
 
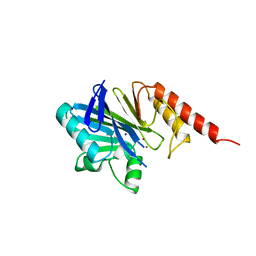 | | METALLO-BETA-LACTAMASE | | Descriptor: | METALLO-BETA-LACTAMASE, SODIUM ION, ZINC ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis.
Structure, 4, 1996
|
|
4YZC
 
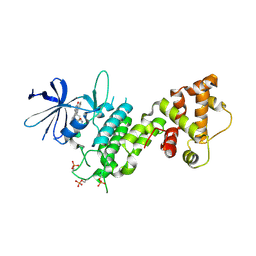 | | Crystal structure of pIRE1alpha in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
4YZ9
 
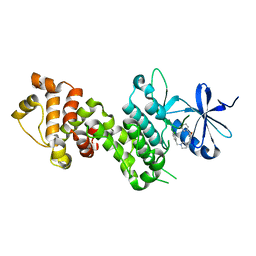 | |
4YZD
 
 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
1GFW
 
 | |
6NT2
 
 | | type 1 PRMT in complex with the inhibitor GSK3368715 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, N~1~-({5-[4,4-bis(ethoxymethyl)cyclohexyl]-1H-pyrazol-4-yl}methyl)-N~1~,N~2~-dimethylethane-1,2-diamine, ... | | Authors: | Concha, N.O. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss.
Cancer Cell, 36, 2019
|
|
8UIF
 
 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2R)-1-[(2S)-oxolan-2-yl]-3-[(3S)-2-oxooxolan-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UIA
 
 | |
8UHO
 
 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-[(2S,3Z)-1-[(2S)-oxolan-2-yl]-3-(2-oxooxolan-3-ylidene)propan-2-yl]-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
2FVC
 
 | | Crystal structure of NS5B BK strain (delta 24) in complex with a 3-(1,1-Dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinone | | Descriptor: | 3-(1,1-dioxido-4H-1,2,4-benzothiadiazin-3-yl)-4-hydroxy-1-(3-methylbutyl)quinolin-2(1H)-one, polyprotein | | Authors: | Concha, N.O, Wonacott, A, Singh, O. | | Deposit date: | 2006-01-30 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase.
J.Med.Chem., 49, 2006
|
|
3E88
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8C
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor peptide | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E87
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta peptide, N-[(1S)-2-amino-1-phenylethyl]-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)thiophene-2-carboxamide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8D
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8E
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, PKI inhibitor peptide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HHK
 
 | | HCV NS5b polymerase complex with a substituted benzothiadizine | | Descriptor: | 2-({(3R)-3-[(3S)-1-(3-methylbutyl)-2,4-dioxo-1,2,3,4-tetrahydroquinolin-3-yl]-1,1-dioxido-3,4-dihydro-2H-1,2,4-benzothiadiazin-7-yl}oxy)acetamide, HCV NS5 polymerase | | Authors: | Concha, N.O, Singh, O. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted benzothiadizine inhibitors of Hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3D0E
 
 | | Crystal structure of human Akt2 in complex with GSK690693 | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3-yn-2-ol, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Smallwood, A. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase.
J.Med.Chem., 51, 2008
|
|
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
8ULD
 
 | | SARA CoV-2 3C-like protease in complex with GSK3487016A | | Descriptor: | 1,2-ETHANEDIOL, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2S,3R)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]-4-[(propan-2-yl)amino]butan-2-yl}-L-leucinamide, Replicase polyprotein 1a | | Authors: | Williams, S.P, Concha, N.O. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
1NVR
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
