1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
1IZ6
 
 | | Crystal Structure of Translation Initiation Factor 5A from Pyrococcus Horikoshii | | Descriptor: | Initiation Factor 5A | | Authors: | Yao, M, Ohsawa, A, Kikukawa, S, Tanaka, I, Kimura, M. | | Deposit date: | 2002-09-25 | | Release date: | 2003-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hyperthermophilic Archaeal Initiation Factor 5A: A Homologue of Eukaryotic Initiation Factor 5A (eIF-5A)
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1WLS
 
 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ZQ0
 
 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
1V7L
 
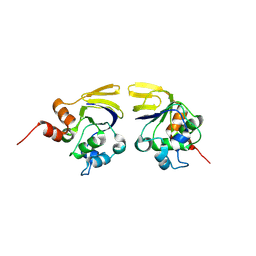 | |
1VGJ
 
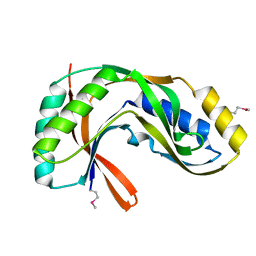 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
4WLC
 
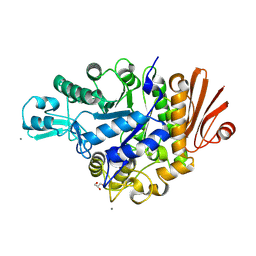 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
4XB3
 
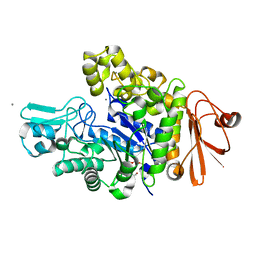 | | Structure of dextran glucosidase | | Descriptor: | CALCIUM ION, Glucan 1,6-alpha-glucosidase, HEXAETHYLENE GLYCOL | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
1EL1
 
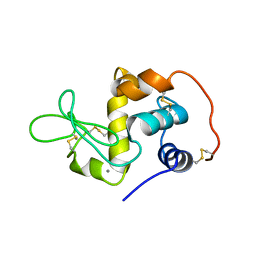 | |
4WJ3
 
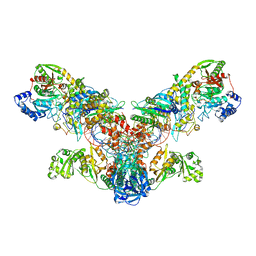 | | Crystal structure of the asparagine transamidosome from Pseudomonas aeruginosa | | Descriptor: | 76mer-tRNA, Aspartate--tRNA(Asp/Asn) ligase, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.705 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WJ4
 
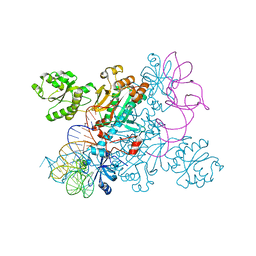 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XD9
 
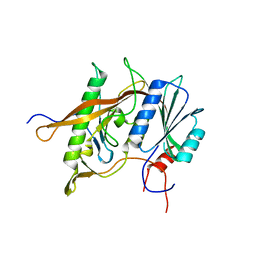 | | Structure of Rpf2-Rrs1 complex involved in ribosome biogenesis | | Descriptor: | Ribosome biogenesis protein (Rrs1), putative (AFU_orthologue AFUA_7G04430), Ribosome biogenesis protein, ... | | Authors: | Asano, N, Kato, K, Yao, M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
1GD6
 
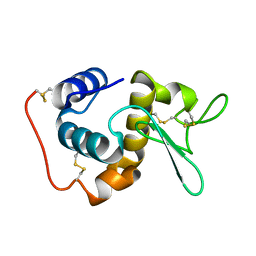 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
1J1G
 
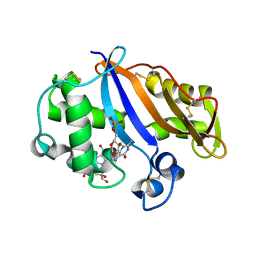 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
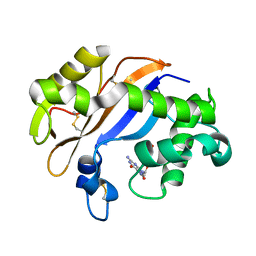 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
5E1Q
 
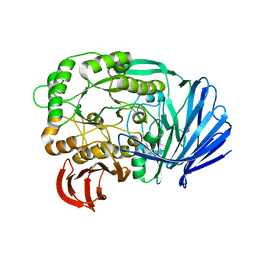 | | Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac | | Descriptor: | CALCIUM ION, GLYCEROL, Retaining alpha-galactosidase, ... | | Authors: | Matsunaga, K, Yamashita, K, Tagami, T, Yao, M, Okuyama, M, Kimura, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
FEBS J., 284, 2017
|
|
5GHA
 
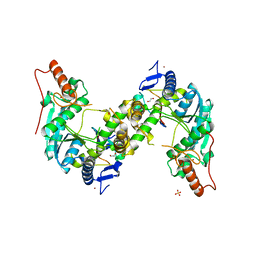 | | Sulfur Transferase TtuA in complex with Sulfur Carrier TtuB | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Sulfur Carrier TtuB, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-06-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1UCA
 
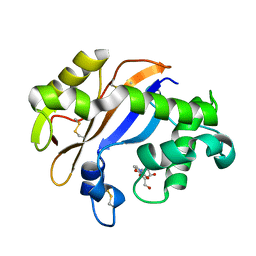 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1QQY
 
 | |
1J0D
 
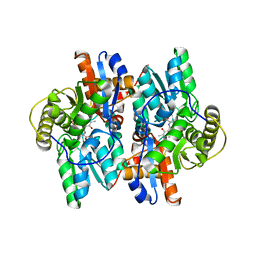 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J1W
 
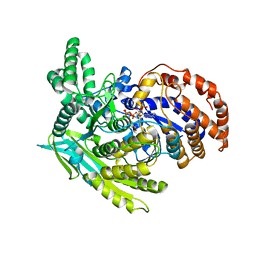 | | Crystal Structure Of The Monomeric Isocitrate Dehydrogenase In Complex With NADP+ | | Descriptor: | Isocitrate Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-12-19 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Monomeric Isocitrate Dehydrogenase in the Presence of NADP+
J.Biol.Chem., 278, 2003
|
|
1J0E
 
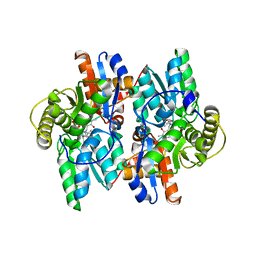 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
4DWQ
 
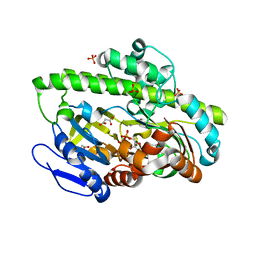 | | RNA ligase RtcB-GMP/Mn(2+) complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MALONATE ION, ... | | Authors: | Okada, C, Xia, S, Englert, M, Yao, M, Soll, D, Wang, J. | | Deposit date: | 2012-02-26 | | Release date: | 2012-09-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OCZ
 
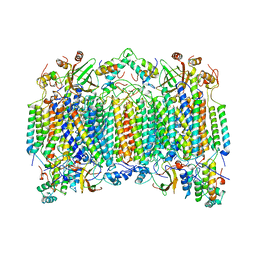 | |
