2AIO
 
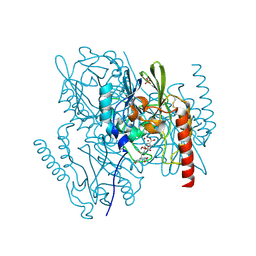 | | Metallo beta lactamase L1 from Stenotrophomonas maltophilia complexed with hydrolyzed moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Spencer, J, Read, J, Sessions, R.B, Howell, S, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2005-07-30 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibiotic Recognition by Binuclear Metallo-beta-Lactamases Revealed by X-ray Crystallography
J.Am.Chem.Soc., 127, 2005
|
|
3IT5
 
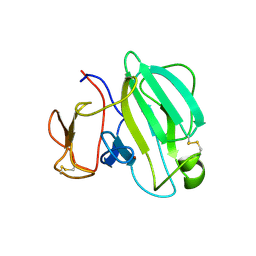 | | Crystal Structure of the LasA Virulence Factor from Pseudomonas aeruginosa | | Descriptor: | Protease lasA, ZINC ION | | Authors: | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
3IT7
 
 | | Crystal Structure of the LasA virulence factor from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Protease lasA, ... | | Authors: | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
4UA4
 
 | |
2QIN
 
 | |
5EV6
 
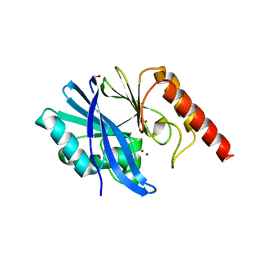 | | Crystal structure of the native, di-zinc metallo-beta-lactamase IMP-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase IMP-1, ... | | Authors: | Spencer, J, Hinchliffe, P. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4WD6
 
 | |
7O0O
 
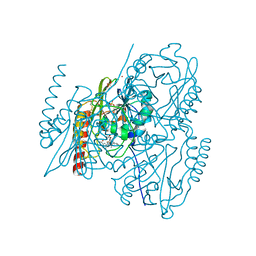 | | Crystal structure of the B3 metallo-beta-lactamase L1 with hydrolysed ertapenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallography and QM/MM Simulations Identify Preferential Binding of Hydrolyzed Carbapenem and Penem Antibiotics to the L1 Metallo-beta-Lactamase in the Imine Form.
J.Chem.Inf.Model., 61, 2021
|
|
2Y87
 
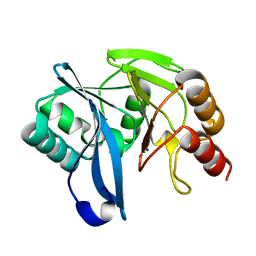 | | Native VIM-7. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
2YNT
 
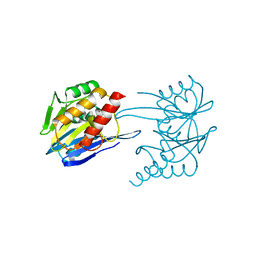 | | GIM-1-3Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
2YNW
 
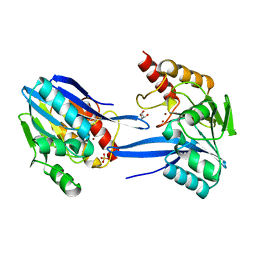 | | GIM-1-2Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, SULFATE ION, ... | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
8BW4
 
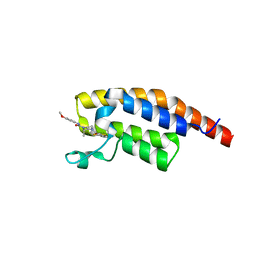 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW2
 
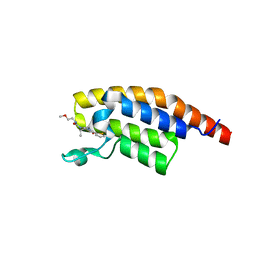 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-N-(2-methoxyethyl)-2-methyl-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
2Y8B
 
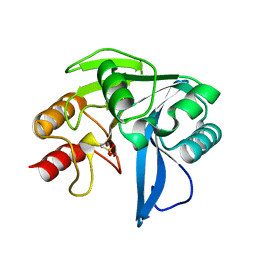 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
7AFZ
 
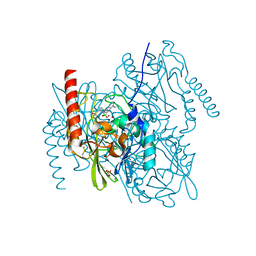 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
2YNV
 
 | | Cys221 oxidized, Mono zinc GIM-1 - GIM-1-Ox. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta- lactamases | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
2Y8A
 
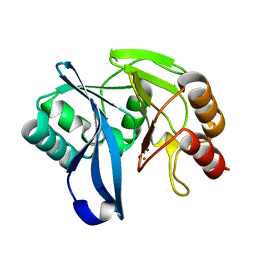 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
2YNU
 
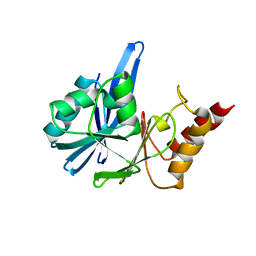 | | Apo GIM-1 with 2Mol. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
3Q6V
 
 | | Crystal Structure of Serratia fonticola Sfh-I: glycerol complex | | Descriptor: | Beta-lactamase, GLYCEROL, ZINC ION | | Authors: | Fonseca, F, Saavedra, M.J, Correia, A, Spencer, J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of Serratia fonticola Sfh-I: activation of the nucleophile in mono-zinc metallo-beta-lactamases.
J.Mol.Biol., 411, 2011
|
|
2QJS
 
 | |
7BH4
 
 | | XFEL structure of apo CTX-M-15 after mixing for 0.7 sec with ertapenem using a piezoelectric injector (PolyPico) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH5
 
 | | XFEL structure of the ertapenem-derived CTX-M-15 acylenzyme after mixing for 2 sec using a piezoelectric injector (PolyPico) | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH7
 
 | | Room temperature, serial X-ray structure of the ertapenem-derived acylenzyme of CTX-M-15 (10 min soak) collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
