3NMH
 
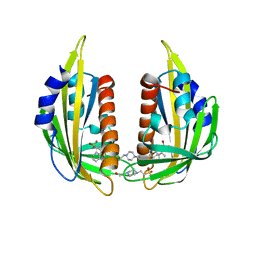 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMV
 
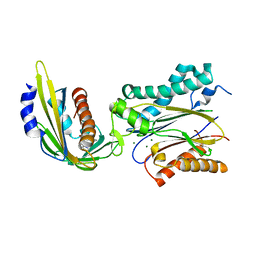 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase ABI2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMN
 
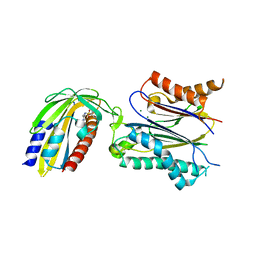 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMP
 
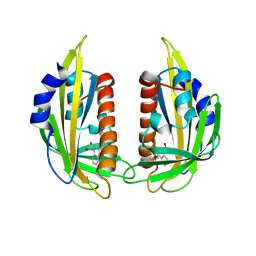 | | Crystal structure of the abscisic receptor PYL2 mutant A93F in complex with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMT
 
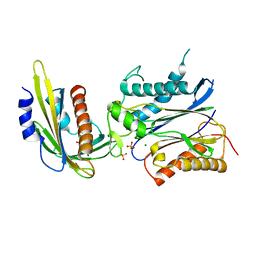 | | Crystal structure of pyrabactin bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase HAB1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KAY
 
 | | Crystal structure of abscisic acid receptor PYL1 | | Descriptor: | Putative uncharacterized protein At5g46790 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB0
 
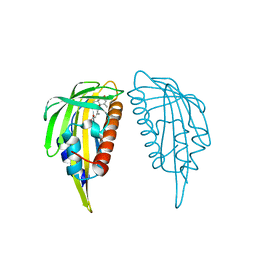 | | Crystal structure of abscisic acid-bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KAZ
 
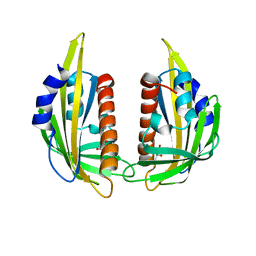 | | Crystal structure of abscisic acid receptor PYL2 | | Descriptor: | 1,3-BUTANEDIOL, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB3
 
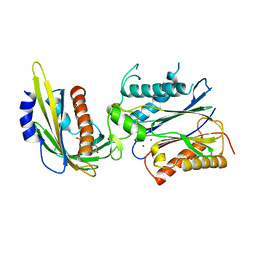 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
6MS7
 
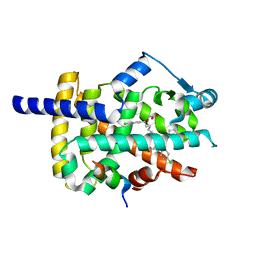 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
7M74
 
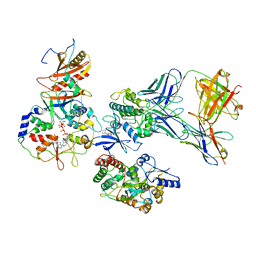 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7S3G
 
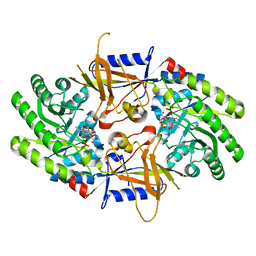 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3F
 
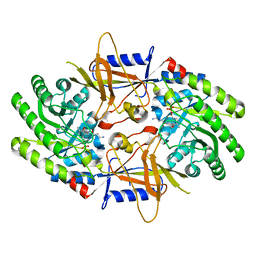 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7U6U
 
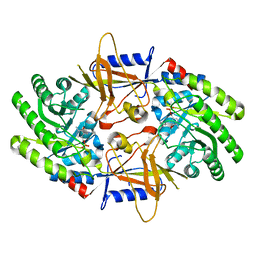 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6P
 
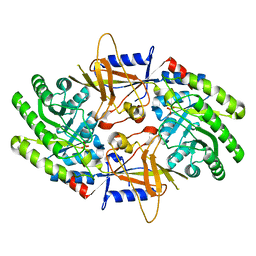 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
5JA5
 
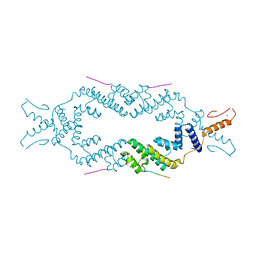 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JHP
 
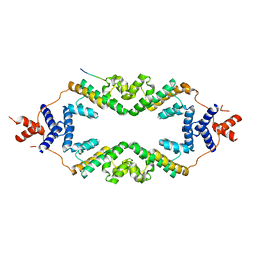 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5J9K
 
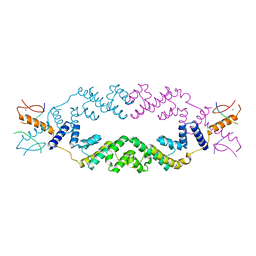 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, ZINC ION, rice D53 peptide 794-808 | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JGC
 
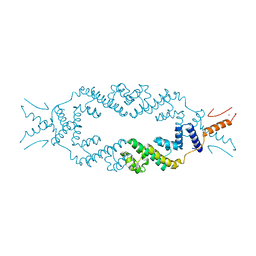 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A, L130A, L179A and I195A mutant | | Descriptor: | Protein TPR1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-20 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5T0Q
 
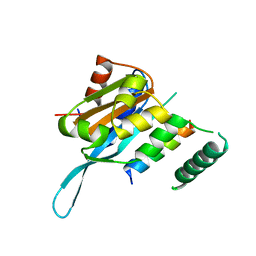 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
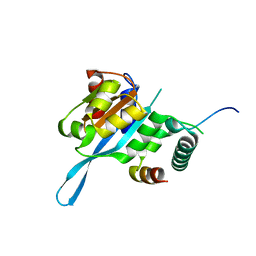 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4RED
 
 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4RER
 
 | | Crystal structure of the phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex bound to AMP and cyclodextrin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.047 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
