1IPA
 
 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1R5Z
 
 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2D00
 
 | | Subunit F of V-type ATPase/synthase | | Descriptor: | CALCIUM ION, V-type ATP synthase subunit F | | Authors: | Makyio, H, Iino, R, Ikeda, C, Imamura, H, Tamakoshi, M, Iwata, M, Stock, D, Bernal, R.A, Carpenter, E.P, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2005-07-21 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
Embo J., 24, 2005
|
|
1WST
 
 | | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, multiple substrate aminotransferase | | Authors: | Lee, W.C, Manabe, F, Nemoto, N, Tamakoshi, M, Tanokura, M, Yamagishi, A. | | Deposit date: | 2004-11-10 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus
To be Published
|
|
2VPX
 
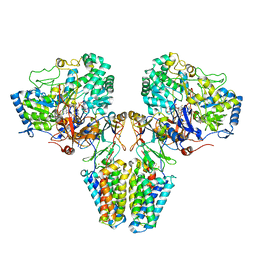 | | Polysulfide reductase with bound quinone (UQ1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPY
 
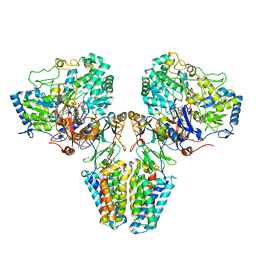 | | Polysulfide reductase with bound quinone inhibitor, pentachlorophenol (PCP) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPW
 
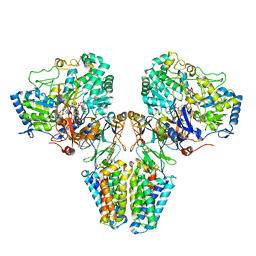 | | Polysulfide reductase with bound menaquinone | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPZ
 
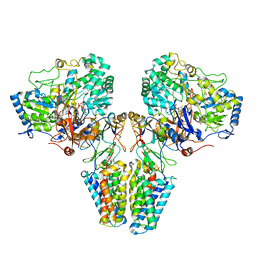 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
6LY9
 
 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6LY8
 
 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
5Y60
 
 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y5Y
 
 | | V/A-type ATPase/synthase from Thermus thermophilus, peripheral domain, rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-24 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y5X
 
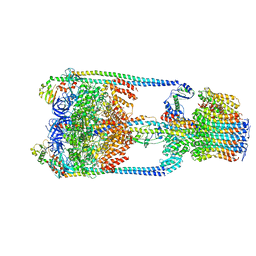 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y5Z
 
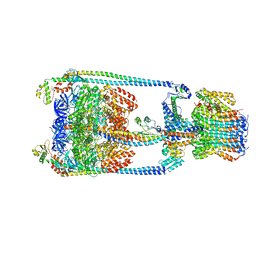 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
2ZDJ
 
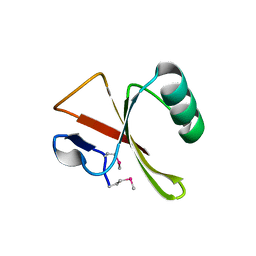 | | Crystal Structure of TTMA177, a Hypothetical Protein from Thermus thermophilus phage TMA | | Descriptor: | hypothetical protein TTMA177 | | Authors: | Agari, Y, Tamakoshi, M, Yamagishi, A, Shinkai, A, Ebihara, A, Yokoyama, S, Kuramitsu, S, Oshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of TTMA177, a Hypothetical Protein from Thermus thermophilus phage TMA
To be Published
|
|
1UIR
 
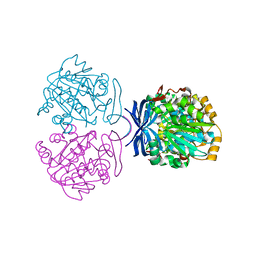 | | Crystal Structure of Polyamine Aminopropyltransfease from Thermus thermophilus | | Descriptor: | Polyamine Aminopropyltransferase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Kumasaka, T, Oshima, T, Tanaka, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
3ANX
 
 | | Crystal structure of triamine/agmatine aminopropyltransferase (SPEE) from thermus thermophilus, complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, spermidine synthase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Tanaka, N, Oshima, T, Kumasaka, T. | | Deposit date: | 2010-09-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
