7ZI0
 
 | | Structure of human Smoothened in complex with cholesterol and SAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, ... | | Authors: | Byrne, E.F.X, Woolley, R.E, Ansell, B, Sansom, M.S.P, Newstead, S, Siebold, C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain.
Sci Adv, 8, 2022
|
|
5FTT
 
 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
5FTU
 
 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7A46
 
 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZYE
 
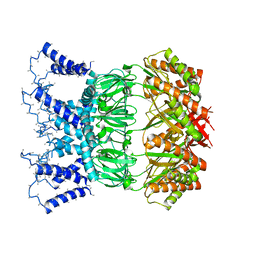 | | YnaI in an open-like conformation | | Descriptor: | YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZYD
 
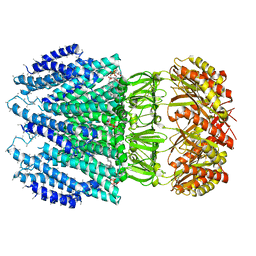 | | YnaI | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2X79
 
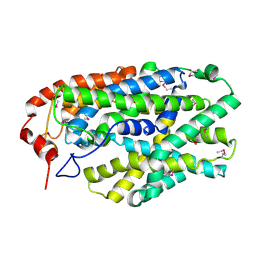 | | Inward facing conformation of Mhp1 | | Descriptor: | HYDANTOIN TRANSPORT PROTEIN | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
2XUT
 
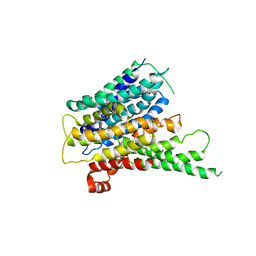 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
6RTW
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain in complex with nanobody NB64 and cholesterol-hemisuccinate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Llama-derived nanobody NB64, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RVC
 
 | | Crystal structure of Patched-1 ectodomain 2 (PTCH1-ECD2) in complex with nanobody 75 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody NB75, Protein patched homolog 1, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RTY
 
 | | Crystal structure of the Patched ectodomain in complex with nanobody NB64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Llama-derived nanobody NB64, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RTX
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RVD
 
 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
3ZRS
 
 | | X-ray crystal structure of a KirBac potassium channel highlights a mechanism of channel opening at the bundle-crossing gate. | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION | | Authors: | Bavro, V.N, De Zorzi, R, Schmidt, M.R, Muniz, J.R.C, Zubcevic, L, Sansom, M.S.P, Venien-Bryan, C, Tucker, S.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a Kirbac Potassium Channel with an Open Bundle Crossing Indicates a Mechanism of Channel Gating
Nat.Struct.Mol.Biol., 19, 2012
|
|
4BBK
 
 | | Structural and functional characterisation of the kindlin-1 pleckstrin homology domain | | Descriptor: | FERMITIN FAMILY HOMOLOG 1, GLYCEROL | | Authors: | Yates, L.A, Lumb, C.N, Brahme, N.N, Zalyte, R, Bird, L.E, De Colibus, L, Owens, R.J, Calderwood, D.A, Sansom, M.S.P, Gilbert, R.J.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterisation of the Kindlin-1 Pleckstrin Homology Domain
J.Biol.Chem., 287, 2012
|
|
8C5V
 
 | |
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
6M96
 
 | |
6NF4
 
 | | Structure of zebrafish Otop1 in nanodiscs | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, Otopetrin1 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NF6
 
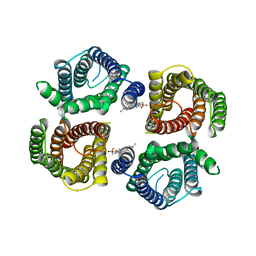 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7Q6Z
 
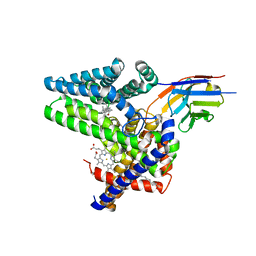 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | | Descriptor: | 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl]ethanone, CHOLESTEROL, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
7Q1U
 
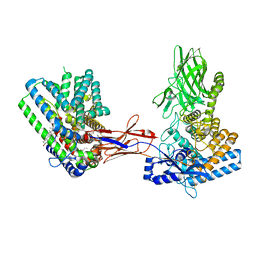 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map) | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
7M6S
 
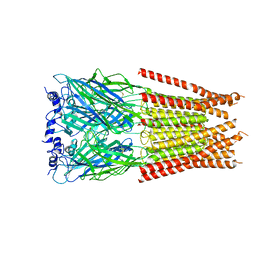 | |
7M6O
 
 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6P
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
