1XDK
 
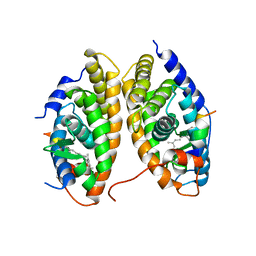 | | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator | | Descriptor: | (9cis)-retinoic acid, Retinoic acid receptor RXR-alpha, Retinoic acid receptor, ... | | Authors: | Pogenberg, V, Guichou, J.F, Vivat-Hannah, V, Kammerer, S, Perez, E, Germain, P, De Lera, A.R, Gronemeyer, H, Royer, C.A, Bourguet, W. | | Deposit date: | 2004-09-07 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CHARACTERIZATION OF THE INTERACTION BETWEEN RAR/RXR HETERODIMERS AND TRANSCRIPTIONAL COACTIVATORS THROUGH STRUCTURAL AND FLUORESCENCE ANISOTROPY STUDIES
J.Biol.Chem., 280, 2005
|
|
4ATI
 
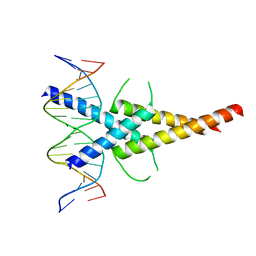 | | MITF:M-box complex | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*CP*AP*TP*GP*TP*GP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*CP*AP*CP*AP*TP*GP*AP*CP*CP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
4ATK
 
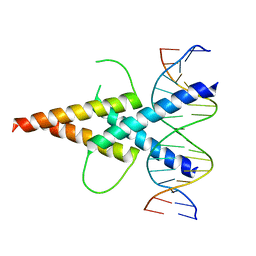 | | MITF:E-box complex | | Descriptor: | 5'-D(*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
4ATH
 
 | | MITF apo structure | | Descriptor: | MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
6FX5
 
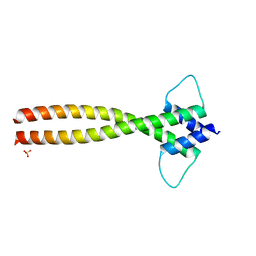 | | MITF dimerization mutant | | Descriptor: | Microphthalmia-associated transcription factor, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of conditional partner selectivity in MITF/TFE family transcription factors with a conserved coiled coil stammer motif.
Nucleic Acids Res., 48, 2020
|
|
2WT7
 
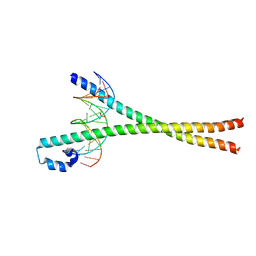 | | Crystal structure of the bZIP heterodimeric complex MafB:cFos bound to DNA | | Descriptor: | MODIFIED T-MARE MOTIF, PHOSPHATE ION, PROTO-ONCOGENE PROTEIN C-FOS, ... | | Authors: | Pogenberg, V, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-11 | | Release date: | 2010-09-29 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
6G1L
 
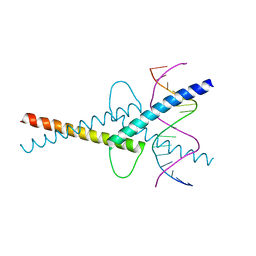 | | MITF/CLEARbox structure | | Descriptor: | CLEAR-box, Microphthalmia-associated transcription factor | | Authors: | Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-03-21 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MITF has a central role in regulating starvation-induced autophagy in melanoma.
Sci Rep, 9, 2019
|
|
2XIV
 
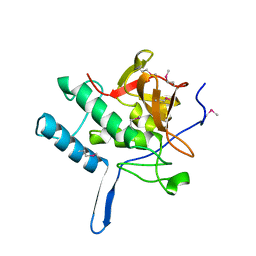 | |
3L1P
 
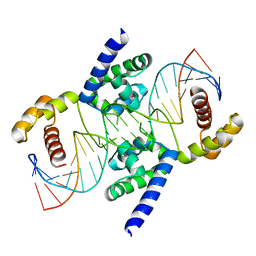 | | POU protein:DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP*TP*CP*AP*AP*AP*TP*GP*TP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP*CP*AP*AP*AP*TP*GP*GP*A)-3'), POU domain, ... | | Authors: | Vahokoski, J, Groves, M.R, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2009-12-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique Oct4 interface is crucial for reprogramming to pluripotency
Nat.Cell Biol., 15, 2013
|
|
6HT5
 
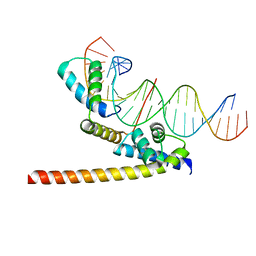 | | Oct4/Sox2:UTF1 structure | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*AP*GP*CP*AP*TP*AP*AP*CP*AP*AP*TP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*CP*AP*TP*TP*GP*TP*TP*AP*TP*GP*CP*TP*AP*GP*TP*GP*AP*AP*G)-3'), POU domain, ... | | Authors: | Vahokoski, J, Meusch, D, Groves, M, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | Oct4/Sox2:UTF1 structure
To Be Published
|
|
6ZMD
 
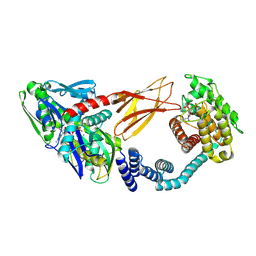 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
6SIU
 
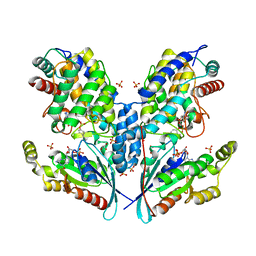 | | Crystal structure of IbpAFic2 covalently tethered to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gulen, B, Roselin, M, Albers, M, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture.
Nat.Chem., 12, 2020
|
|
3ZMP
 
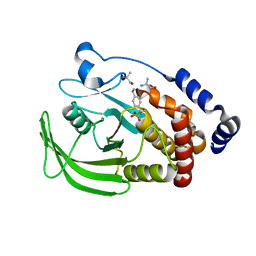 | | Src-derived peptide inhibitor complex of PTP1B | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Temmerman, K, Pogenberg, V, Meyer, C, Koehn, M, Wilmanns, M. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Development of Accessible Peptidic Tool Compounds to Study the Phosphatase Ptp1B in Intact Cells.
Acs Chem.Biol., 9, 2014
|
|
3ZMQ
 
 | | Src-derived mutant peptide inhibitor complex of PTP1B | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Temmerman, K, Pogenberg, V, Meyer, C, Koehn, M, Wilmanns, M. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development of Accessible Peptidic Tool Compounds to Study the Phosphatase Ptp1B in Intact Cells.
Acs Chem.Biol., 9, 2014
|
|
6FKG
 
 | | Crystal structure of the M.tuberculosis MbcT-MbcA toxin-antitoxin complex. | | Descriptor: | GLYCEROL, Rv1989c (MbcT), Rv1990c (MbcA) | | Authors: | Freire, D.M, Cianci, M, Pogenberg, V, Schneider, T.R, Wilmanns, M, Parret, A.H.A. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An NAD+Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol.Cell, 73, 2019
|
|
8AGG
 
 | |
8ALK
 
 | | Structure of the Legionella phosphocholine hydrolase Lem3 in complex with its substrate Rab1 | | Descriptor: | 2-[[[5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxyethyl-[3-(2-chloranylethanoylamino)propyl]-dimethyl-azanium, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kaspers, M.S, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
8ANP
 
 | | Legionella effector Lem3 mutant D190A in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Phosphocholine hydrolase Lem3, SULFATE ION, ... | | Authors: | Kaspers, M.S, Pogenberg, V, Ernst, S, Ecker, F, Pett, C, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
8CIL
 
 | |
4B4L
 
 | | CRYSTAL STRUCTURE OF AN ARD DAP-KINASE 1 MUTANT | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Temmerman, K, Pogenberg, V, Jonko, W, Wilmanns, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
4CHG
 
 | | Crystal structure of VapBC15 complex from Mycobacterium tuberculosis | | Descriptor: | ANTITOXIN VAPB15, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Das, U, Pogenberg, V, Tiruttani Subhramanyam, U.K, Wilmanns, M, Srinivasan, A, Gourinath, S. | | Deposit date: | 2013-12-02 | | Release date: | 2014-11-12 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Vapbc-15 Complex from Mycobacterium Tuberculosis Reveals a Two-Metal Ion Dependent Pin-Domain Ribonuclease And a Variable Mode of Toxin-Antitoxin Assembly.
J.Struct.Biol., 188, 2014
|
|
2WTY
 
 | | Crystal structure of the homodimeric MafB in complex with the T-MARE binding site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP *GP*CP*AP*AP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*TP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP *GP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Consani Textor, L, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-25 | | Release date: | 2010-12-08 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
5AF3
 
 | |
5NEW
 
 | |
3MZH
 
 | |
