5HLO
 
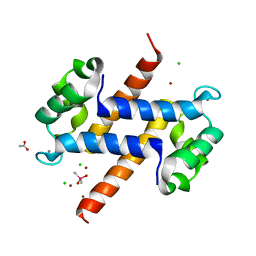 | | Crystal structure of calcium and zinc-bound human S100A8 in space group C2221 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
5HLV
 
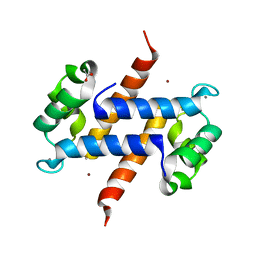 | | Crystal structure of calcium and zinc-bound human S100A8 in space group P212121 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
7XKE
 
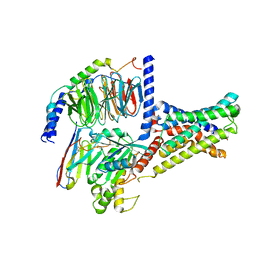 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
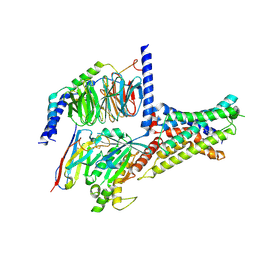 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
6IS8
 
 | | Crystal structure of ZmMoc1 D115N mutant in complex with Holliday junction | | Descriptor: | DNA (33-MER), MAGNESIUM ION, Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6IS9
 
 | | Crystal Structure of ZmMOC1 | | Descriptor: | Monokaryotic chloroplast 1 | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JRG
 
 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JRF
 
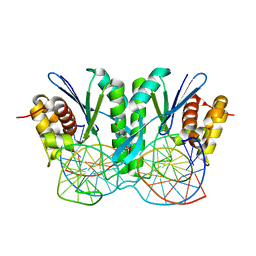 | | Crystal structure of ZmMoc1-Holliday junction Complex in the presence of Calcium | | Descriptor: | CALCIUM ION, DNA (33-MER), Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6K1S
 
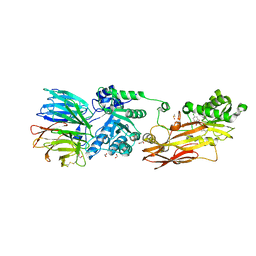 | | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase (PRMT5) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[7-[(2~{R},3~{R},4~{S},5~{R})-5-[(~{R})-(4-chlorophenyl)-oxidanyl-methyl]-3,4-bis(oxidanyl)oxolan-2-yl]pyrrolo[2,3-d]pyrimidin-4-yl]amino]ethanal, DIMETHYL SULFOXIDE, ... | | Authors: | Tong, S, Lin, H. | | Deposit date: | 2019-05-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
8TMR
 
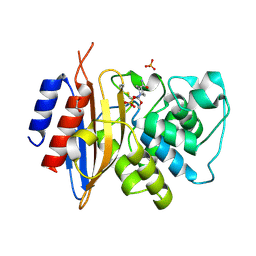 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMT
 
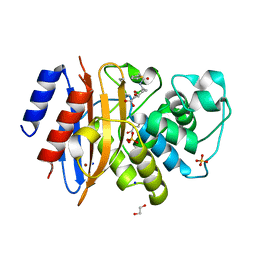 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TN0
 
 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
3ZJ6
 
 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
7RK3
 
 | |
7ROM
 
 | |
3ZJ8
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | Descriptor: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3I9N
 
 | | Crystal structure of human CD38 complexed with an analog ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9K
 
 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9M
 
 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9J
 
 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9L
 
 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
4EXP
 
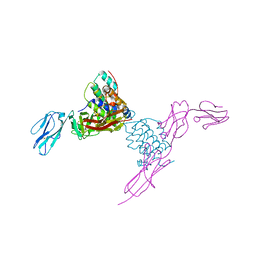 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
