4U6D
 
 | | Zg3615, a family 117 glycoside hydrolase in complex with beta-3,6-anhydro-L-galactose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-beta-L-galactopyranose, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U6B
 
 | | Zg3597, a family 117 glycoside hydrolase, produced by the marine bacterium Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3REN
 
 | | CPF_2247, a novel alpha-amylase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase, family 8, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Boraston, A.B. | | Deposit date: | 2011-04-04 | | Release date: | 2011-05-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Proteins, 79, 2011
|
|
5OPQ
 
 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
2J1A
 
 | |
2J1E
 
 | |
2J7M
 
 | | Characterization of a Family 32 CBM | | Descriptor: | CALCIUM ION, HYALURONIDASE, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2006-10-12 | | Release date: | 2006-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Interaction of a Carbohydrate-Binding Module from a Clostridium Perfringens N-Acetyl-Beta-Hexosaminidase with its Carbohydrate Receptor
J.Biol.Chem., 281, 2006
|
|
4A3Z
 
 | | CpGH89CBM32-4 (seleno-methionine labeled) produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A45
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A41
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with galactose | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, SODIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A44
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylglucosaminidase, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4AAX
 
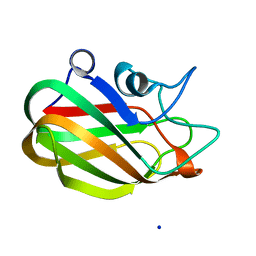 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-12-05 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A42
 
 | | CpGH89CBM32-6 produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A4A
 
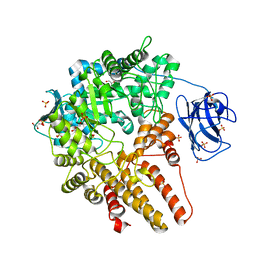 | | CpGH89 (E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-07 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a bacterial exo-alpha-D-N-acetylglucosaminidase in complex with an unusual disaccharide found in class III mucin.
Glycobiology, 22, 2012
|
|
4A6O
 
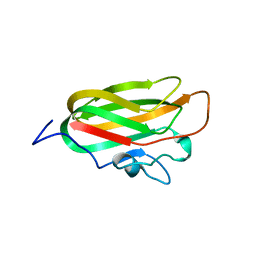 | | CpGH89CBM32-4, produced by Clostridium perfringens, in complex with glcNAc-alpha-1,4-galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
2W1U
 
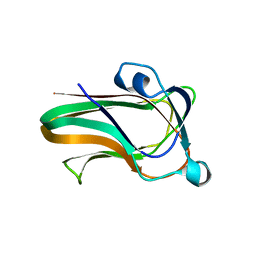 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ACETATE ION, ... | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2W1Q
 
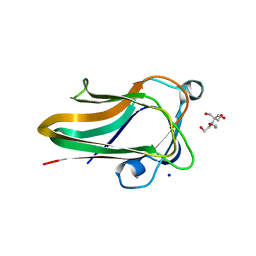 | |
2WDB
 
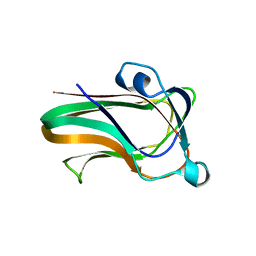 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2W1S
 
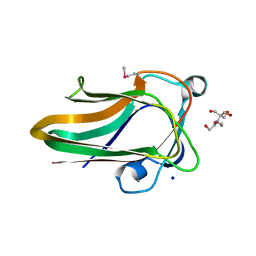 | |
2W1N
 
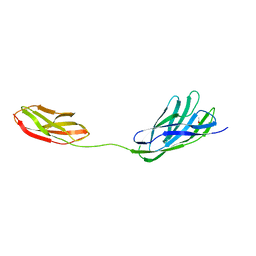 | | cohesin and fibronectin type-III double module construct from the Clostridium perfringens glycoside hydrolase GH84C | | Descriptor: | ACETATE ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Czjzek, M, Smith, S.J, Boraston, A.B. | | Deposit date: | 2008-10-17 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2V5C
 
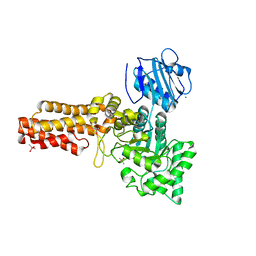 | | Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure | | Descriptor: | CACODYLATE ION, CALCIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2V5D
 
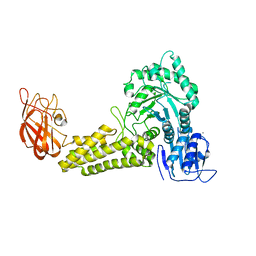 | | Structure of a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. | | Descriptor: | CALCIUM ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2VCA
 
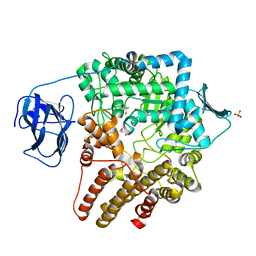 | | Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VCB
 
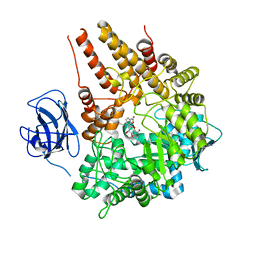 | | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VCC
 
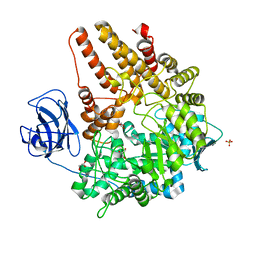 | | Family 89 Glycoside Hydrolase from Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
