8GHI
 
 | |
8GHH
 
 | |
4X9X
 
 | | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid Binding Protein Family | | Descriptor: | 1,2-ETHANEDIOL, DegV domain-containing protein MW1315, OLEIC ACID | | Authors: | Broussard, T.C, Miller, D.J, Jackson, P, Nourse, A, Rock, C.O. | | Deposit date: | 2014-12-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid-binding Protein Family.
J.Biol.Chem., 291, 2016
|
|
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
1EBL
 
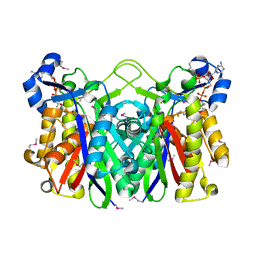 | | THE 1.8 A CRYSTAL STRUCTURE AND ACTIVE SITE ARCHITECTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE III (FABH) FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL-ACP SYNTHASE III, COENZYME A | | Authors: | Davies, C, Heath, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.
Structure Fold.Des., 8, 2000
|
|
8VDA
 
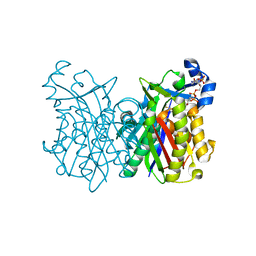 | |
8VD9
 
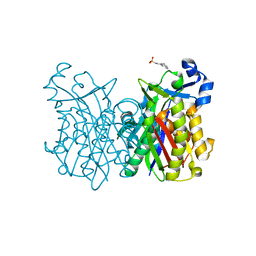 | |
8VDB
 
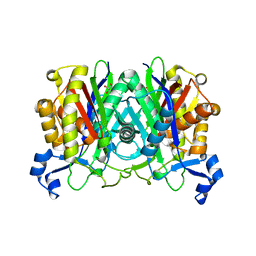 | |
1U1Z
 
 | | The Structure of (3R)-hydroxyacyl-ACP dehydratase (FabZ) | | Descriptor: | (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase, SULFATE ION | | Authors: | Kimber, M.S, Martin, F, Lu, Y, Houston, S, Vedadi, M, Dharamsi, A, Fiebig, K.M, Schmid, M, Rock, C.O. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
1ESM
 
 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | COENZYME A, PANTOTHENATE KINASE | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
1ESN
 
 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | MAGNESIUM ION, PANTOTHENATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
1FJ8
 
 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH CERULENIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1FJ4
 
 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1G5X
 
 | | The Structure of Beta-Ketoacyl-[Acyl Carrier Protein] Synthase I | | Descriptor: | BETA-KETOACYL ACYL CARRIER PROTEIN SYNTHASE I | | Authors: | Zhang, Y.M, Rao, M.S, Heath, R.J, Price, A.C, Olson, A.J, Rock, C.O, White, S.W. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III.
J.Biol.Chem., 276, 2001
|
|
1HW1
 
 | | THE FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ESCHERICHIA COLI | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION, ZINC ION | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1HW2
 
 | | FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ECHERICHIA COLI | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*GP*TP*CP*CP*GP*AP*CP*CP*AP*GP*AP*TP*GP*CP*T)-3', 5'-D(*G*CP*AP*TP*CP*TP*GP*GP*TP*CP*GP*GP*AP*CP*CP*AP*GP*AP*TP*CP*GP*A)-3', FATTY ACID METABOLISM REGULATOR PROTEIN, ... | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1I01
 
 | |
7KAY
 
 | |
7KAX
 
 | |
7KAV
 
 | |
7KAZ
 
 | |
7KAW
 
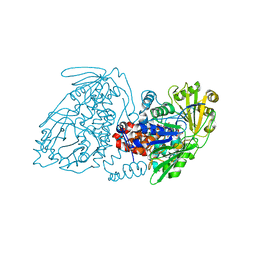 | |
2ALM
 
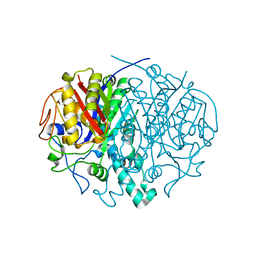 | | Crystal structure analysis of a mutant beta-ketoacyl-[acyl carrier protein] synthase II from Streptococcus pneumoniae | | Descriptor: | 3-oxoacyl-(acyl-carrier-protein) synthase II, MAGNESIUM ION | | Authors: | Zhang, Y.M, Hurlbert, J, White, S.W, Rock, C.O. | | Deposit date: | 2005-08-07 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae.
J.Biol.Chem., 281, 2006
|
|
2F9T
 
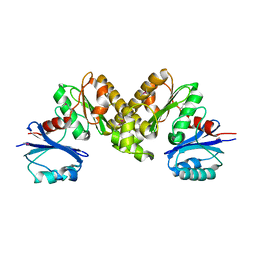 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | Pantothenate Kinase | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
2F9W
 
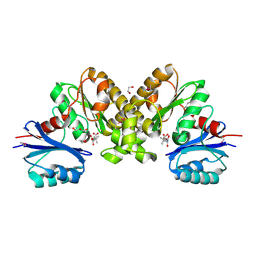 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
