1SIS
 
 | | SPATIAL STRUCTURE OF INSECTOTOXIN I5A BUTHUS EUPEUS BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY (RUSSIAN) | | Descriptor: | SCORPION INSECTOTOXIN I5A | | Authors: | Arseniev, A.S, Kondakov, V.I, Lomize, A.L, Maiorov, V.N, Bystrov, V.F. | | Deposit date: | 1993-11-11 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the spatial structure of insectotoxin 15A from Buthus erpeus by (1)H-NMR spectroscopy data
Bioorg.Khim., 17, 1991
|
|
1GRM
 
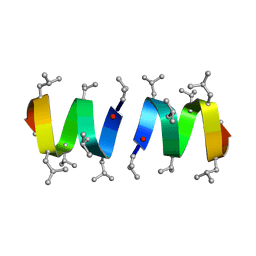 | | REFINEMENT OF THE SPATIAL STRUCTURE OF THE GRAMICIDIN A TRANSMEMBRANE ION-CHANNEL (RUSSIAN) | | Descriptor: | GRAMICIDIN A | | Authors: | Arseniev, A.S, Barsukov, I.L, Lomize, A.L, Orekhov, V.Y, Bystrov, V.F. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Refinement of the Spatial Structure of the Gramicidin a Ion Channel
Biol.Membr.(Ussr), 18, 1992
|
|
1ZAD
 
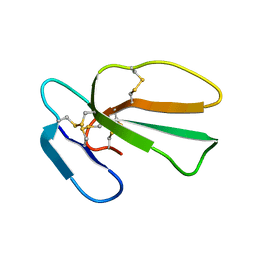 | | Structure of cytotoxin I (CTI) from Naja Oxiana in complex with DPC micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Konshina, A.G, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-06-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins
Biochem.J., 387, 2005
|
|
6HJ7
 
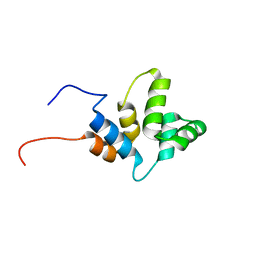 | |
1ZNU
 
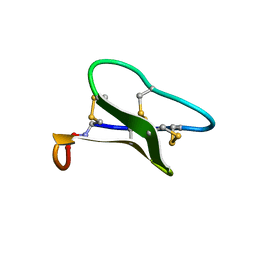 | | Structure of cyclotide Kalata B1 in DPC micelles solution | | Descriptor: | Kalata B1 | | Authors: | Shenkarev, Z.O, Nadezhdin, K.D, Sobol, V.A, Sobol, A.G, Skjeldal, L, Arseniev, A.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-04-25 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle.
Febs J., 273, 2006
|
|
3MRA
 
 | | M3 TRANSMEMBRANE SEGMENT OF ALPHA-SUBUNIT OF NICOTINIC ACETYLCHOLINE RECEPTOR FROM TORPEDO CALIFORNICA, NMR, 15 STRUCTURES | | Descriptor: | Acetylcholine receptor subunit alpha | | Authors: | Lugovskoy, A.A, Maslennikov, I.V, Utkin, Y.N, Tsetlin, V.I, Cohen, J.B, Arseniev, A.S. | | Deposit date: | 1997-07-15 | | Release date: | 1998-01-21 | | Last modified: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | Spatial structure of the M3 transmembrane segment of the nicotinic acetylcholine receptor alpha subunit.
Eur.J.Biochem., 255, 1998
|
|
1R9U
 
 | | Refined structure of peptaibol zervamicin IIB in methanol solution from trans-hydrogen bond J couplings | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balashova, T.A, Yakimenko, Z.A, Ovchinnikova, T.V, Arseniev, A.S. | | Deposit date: | 2003-10-31 | | Release date: | 2004-11-09 | | Last modified: | 2018-10-10 | | Method: | SOLUTION NMR | | Cite: | Biosynthetic Uniform 13C,15N-Labelling of Zervamicin Iib. Complete 13C and 15N NMR Assignment.
J.Pept.Sci., 9, 2003
|
|
6GWM
 
 | | Solution structure of rat RIP2 caspase recruitment domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Mineev, K.S, Goncharuk, S.A, Artemieva, L.E, Arseniev, A.S. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | CARD domain of rat RIP2 kinase: Refolding, solution structure, pH-dependent behavior and protein-protein interactions.
PLoS ONE, 13, 2018
|
|
8C5J
 
 | | Spatial structure of Lch-alpha peptide from two-component lantibiotic system Lichenicidin VK21 | | Descriptor: | Lantibiotic lichenicidin VK21 A1 | | Authors: | Mineev, K.S, Paramonov, A.S, Arseniev, A.S, Ovchinnikova, T.V, Shenkarev, Z.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Specific Binding of the alpha-Component of the Lantibiotic Lichenicidin to the Peptidoglycan Precursor Lipid II Predetermines Its Antimicrobial Activity.
Int J Mol Sci, 24, 2023
|
|
1BHB
 
 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
1BHA
 
 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
6YTS
 
 | | Solution NMR structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | Trichobakin | | Authors: | Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Boyko, K.M, Arseniev, A.S, Usanov, S.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
8AR2
 
 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
1FW7
 
 | |
1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
1IH9
 
 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
1J5J
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1RQT
 
 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQV
 
 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RL5
 
 | | NMR structure with tightly bound water molecule of cytotoxin I from Naja oxiana in aqueous solution (major form) | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2003-11-25 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins.
Biochem.J., 387, 2005
|
|
1SCO
 
 | | SCORPION TOXIN (OSK1 TOXIN) WITH HIGH AFFINITY FOR SMALL CONDUCTANCE CA(2+)-ACTIVATED K+ CHANNEL IN NEUROBLASTOMA-X-GLUOMA NG 108-15 HYBRID CELLS, NMR, 30 STRUCTURES | | Descriptor: | SCORPION TOXIN OSK1 | | Authors: | Jaravine, V.A, Nolde, D.E, Pluzhnikov, K.A, Grishin, E.V, Arseniev, A.S. | | Deposit date: | 1996-04-01 | | Release date: | 1997-01-27 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of toxin OSK1 from Orthochirus scrobiculosus scorpion venom.
Biochemistry, 36, 1997
|
|
8AR1
 
 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
