5LI4
 
 | | bacteriophage phi812K1-420 tail sheath protein after contraction | | Descriptor: | tail sheath protein | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-19 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LIJ
 
 | | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Descriptor: | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LI2
 
 | | bacteriophage phi812K1-420 tail sheath and tail tube protein in native tail | | Descriptor: | Phage-like element PBSX protein XkdM, tail sheath protein | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-19 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LII
 
 | | bacteriophage phi812K1-420 major capsid protein | | Descriptor: | Major capsid protein | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5A8F
 
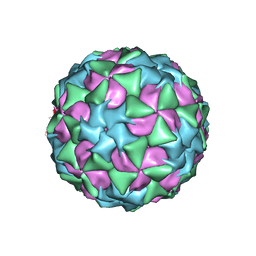 | | Structure and genome release mechanism of human cardiovirus Saffold virus-3 | | Descriptor: | GENOME POLYPHUMAN SAFFOLD VIRUS-3 VP3 PROTEIN, HUMAN SAFFOLD VIRUS-3 VP1, HUMAN SAFFOLD VIRUS-3 VP2 | | Authors: | Mullapudi, E, Novacek, J, Palkova, L, Kulich, P, Lindberg, M, vanKuppeveld, F.J.M, Plevka, P. | | Deposit date: | 2015-07-15 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and Genome Release Mechanism of Human Cardiovirus Saffold Virus-3.
J.Virol., 90, 2016
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
6S47
 
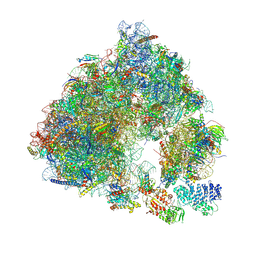 | | Saccharomyces cerevisiae 80S ribosome bound with ABCF protein New1 | | Descriptor: | 18S rRNA (1707-MER), 28S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Kasari, V, Pochopien, A.A, Margus, T, Murina, V, Turnbull, K, Zhou, Y, Nissan, T, Graf, M, Novacek, J, Atkinson, G.C, Johansson, M.J.O, Wilson, D.N, Hauryliuk, V. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A role for the Saccharomyces cerevisiae ABCF protein New1 in translation termination/recycling.
Nucleic Acids Res., 47, 2019
|
|
2KRC
 
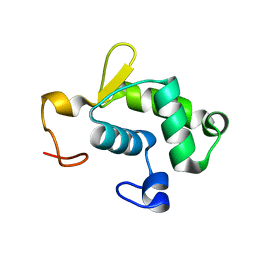 | | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Motackova, V, Sanderova, H, Zidek, L, Novacek, J, Padrta, P, Svenkova, A, Jonak, J, Krasny, L, Sklenar, V. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Proteins, 78, 2010
|
|
5G51
 
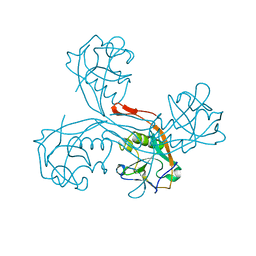 | | High resolution structure of the part of VP3 protein of Deformed Wing Virus forming P-domain | | Descriptor: | DWV-VP3-P-DOMAIN | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-22 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G52
 
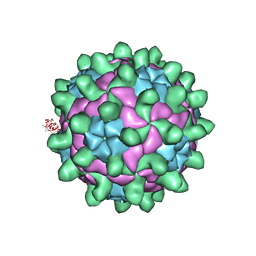 | | Crystallographic structure of full particle of Deformed Wing Virus | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-22 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NWW
 
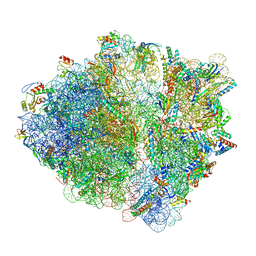 | | CspA-27 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OII
 
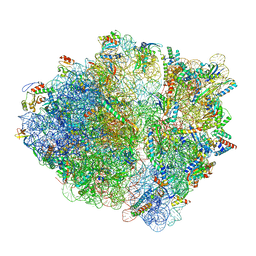 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OT5
 
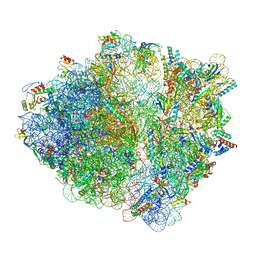 | | CspA-70 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIF
 
 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIG
 
 | | CspA-27 cotranslational folding intermediate 3 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
2M4K
 
 | | Solution structure of the delta subunit of RNA polymerase from Bacillus subtilis | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Papouskova, V, Novacek, J, Kaderavek, P, Zidek, L, Rabatinova, A, Sanderova, H, Krasny, L, Sklenar, V. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Study of the Partially Disordered Full-Length delta Subunit of RNA Polymerase from Bacillus subtilis.
Chembiochem, 14, 2013
|
|
8OOH
 
 | |
8OKA
 
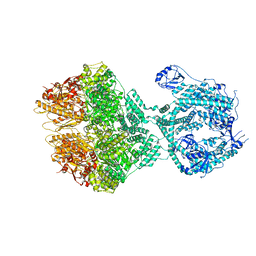 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Phosphorylation of the N-terminal domain of human mitochondrial Lon protease disrupts its functions
To Be Published
|
|
8OM7
 
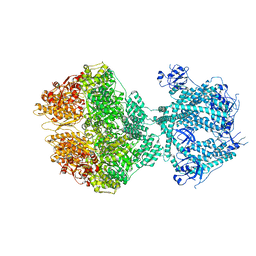 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Phosphorylation of the N-terminal domain of human mitochondrial Lon protease disrupts its functions
To Be Published
|
|
7BC3
 
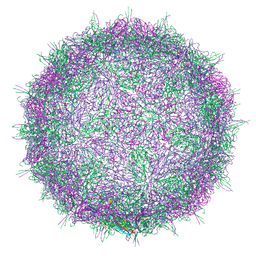 | | Native virion of Kashmir bee virus at acidic pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
7BGK
 
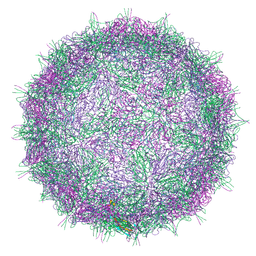 | | Native virion of Kashmir bee virus at neutral pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
7O3D
 
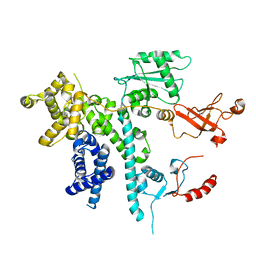 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O6B
 
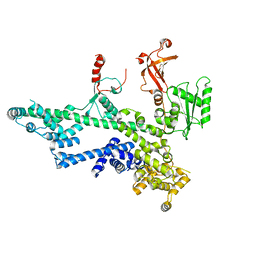 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
6HA8
 
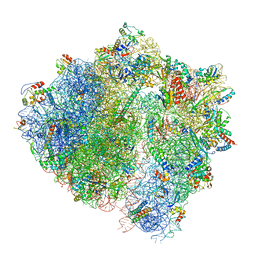 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HXX
 
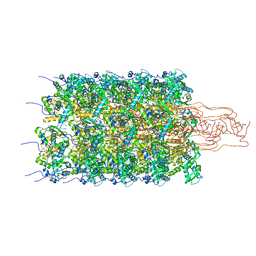 | | Potato virus Y | | Descriptor: | Coat protein, RNA (5'-R(P*UP*UP*UP*UP*U)-3') | | Authors: | Podobnik, M, Kezar, A, Novacek, J, Polak, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the multitasking nature of the potato virus Y coat protein.
Sci Adv, 5, 2019
|
|
