5MKK
 
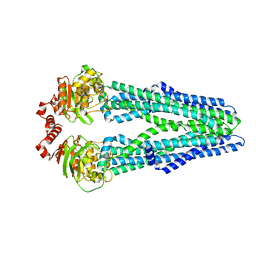 | | Crystal structure of the heterodimeric ABC transporter TmrAB, a homolog of the antigen translocation complex TAP | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, SULFATE ION | | Authors: | Noell, A, Thomas, C, Tomasiak, T.M, Olieric, V, Wang, M, Diederichs, K, Stroud, R.M, Pos, K.M, Tampe, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LIS
 
 | |
5LIN
 
 | |
5LIO
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
