2ZO7
 
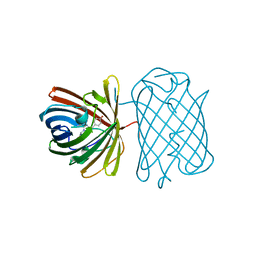 | | Crystal Structure of a Kusabira-Cyan Mutant (KCY-R1), a Cyan/Green-Emitting GFP-Like Protein | | Descriptor: | CYAN/GREEN-EMITTING GFP-LIKE PROTEIN, KUSABIRA-CYAN MUTANT (KCY-R1) | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a new cyan fluorescent protein and its hue-shifted variants
Biochemistry, 48, 2009
|
|
2ZO6
 
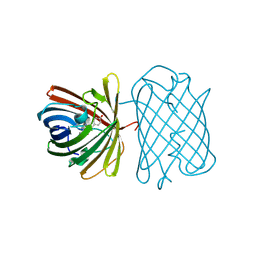 | | Crystal Structure of Kusabira-Cyan (KCY), a Cyan-Emitting GFP-Like Protein | | Descriptor: | CYAN-EMITTING GFP-LIKE PROTEIN, KUSABIRA-CYAN (KCY) | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a new cyan fluorescent protein and its hue-shifted variants
Biochemistry, 48, 2009
|
|
2ZMU
 
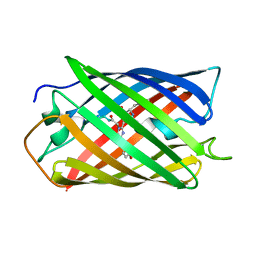 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 9.1 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
2ZMW
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 6.0 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
1GCU
 
 | |
7XI9
 
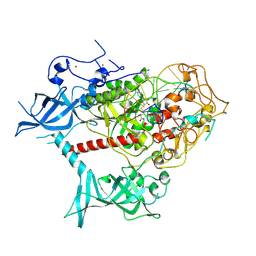 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
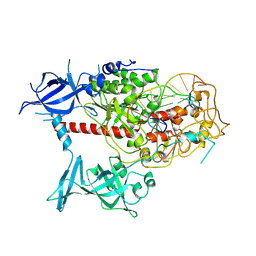 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
2Z1O
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-05-10 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6X
 
 | | Crystal structure of 22G, the wild-type protein of the photoswitchable GFP-like protein Dronpa | | Descriptor: | photochromic protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7C24
 
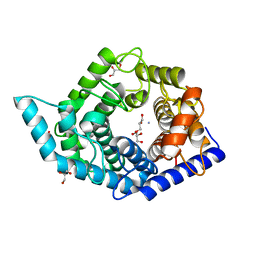 | | Glycosidase F290Y at pH8.0 | | Descriptor: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C25
 
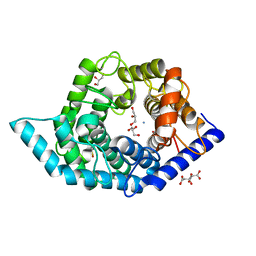 | | Glycosidase Wild Type at pH8.0 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
 | | Glycosidase Wild Type at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C27
 
 | | Glycosidase F290Y at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1WSP
 
 | | Crystal structure of axin dix domain | | Descriptor: | Axin 1 protein, BENZOIC ACID, MERCURY (II) ION | | Authors: | Shibata, N, Hanamura, T, Yamamoto, R, Ueda, Y, Yamamoto, H, Kikuchi, A, Higuchi, Y. | | Deposit date: | 2004-11-08 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of axin dix domain
to be published
|
|
1INZ
 
 | | SOLUTION STRUCTURE OF THE EPSIN N-TERMINAL HOMOLOGY (ENTH) DOMAIN OF HUMAN EPSIN | | Descriptor: | EPS15-INTERACTING PROTEIN(EPSIN) | | Authors: | Koshiba, S, Kigawa, T, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epsin N-terminal homology (ENTH) domain of human epsin
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1IQ3
 
 | | SOLUTION STRUCTURE OF THE EPS15 HOMOLOGY DOMAIN OF A HUMAN POB1 | | Descriptor: | CALCIUM ION, RALBP1-INTERACTING PROTEIN (PARTNER OF RALBP1) | | Authors: | Koshiba, S, Kigawa, T, Iwahara, J, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Eps15 homology domain of a human POB1 (partner of RalBP1).
FEBS Lett., 442, 1999
|
|
5XFM
 
 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
5E1Q
 
 | | Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac | | Descriptor: | CALCIUM ION, GLYCEROL, Retaining alpha-galactosidase, ... | | Authors: | Matsunaga, K, Yamashita, K, Tagami, T, Yao, M, Okuyama, M, Kimura, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
FEBS J., 284, 2017
|
|
5Z3C
 
 | | Glycosidase E178A | | Descriptor: | GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3D
 
 | | Glycosidase F290Y | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3B
 
 | | Glycosidase Y48F | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3A
 
 | | Glycosidase Wild Type | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
