4QBY
 
 | | yCP in complex with BOC-ALA-ALA-ALA-CHO | | Descriptor: | BOC-ALA-ALA-ALA-CHO, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Arciniega, M, Beck, P, Lange, O, Groll, M, Huber, R. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SHJ
 
 | | Proteasome in complex with hydroxyurea derivative HU10 | | Descriptor: | 1-hydroxy-1-[(2R)-4-{3-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yloxy]phenyl}but-3-yn-2-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, ... | | Authors: | Gallastegui, N, Beck, P, Arciniega, M, Hillebrand, S, Huber, R, Groll, M. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hydroxyureas as noncovalent proteasome inhibitors.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3V3V
 
 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
5JXH
 
 | | Structure the proprotein convertase furin in complex with meta-guanidinomethyl-Phac-RVR-Amba at 2.0 Angstrom resolution. | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXJ
 
 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXI
 
 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXG
 
 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F5W
 
 | |
6MAX
 
 | |
4MDN
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
4MDQ
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
6EOP
 
 | | DPP8 - SLRFLYEG, space group 20 | | Descriptor: | CALCIUM ION, CITRATE ANION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOT
 
 | | DPP8 - SLRFLYEG, space group 19 | | Descriptor: | Dipeptidyl peptidase 8, SER-LEU-ARG-PHE-LEU-TYR-GLU-GLY | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOO
 
 | | DPP8 - Apo, space group 20 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOR
 
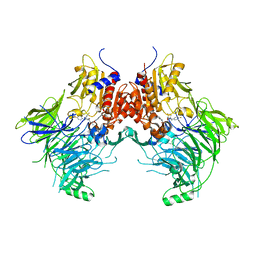 | | DPP9 - 1G244 | | Descriptor: | (2~{S})-2-azanyl-4-[4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-1-(1,3-dihydroisoindol-2-yl)butane-1,4-dione, Dipeptidyl peptidase 9 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOS
 
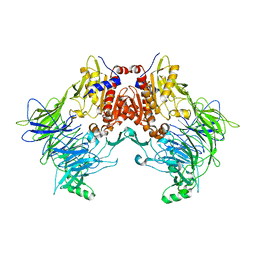 | | DPP8 - Apo, space group 19 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOQ
 
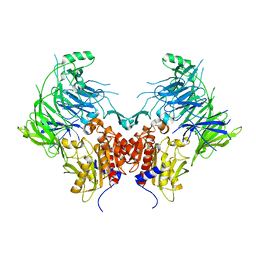 | | DPP9 - Apo | | Descriptor: | Dipeptidyl peptidase 9 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
