1J56
 
 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1LLS
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
1MNL
 
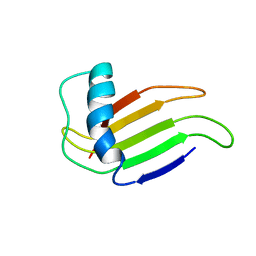 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
7TX7
 
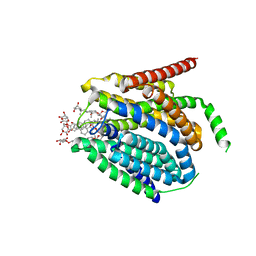 | |
7TX6
 
 | | Cryo-EM structure of the human reduced folate carrier in complex with methotrexate | | Descriptor: | Digitonin, METHOTREXATE, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
6DT0
 
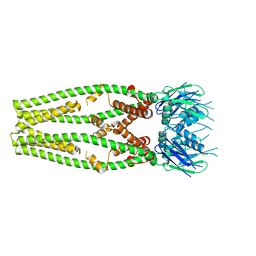 | | Cryo-EM structure of a mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Yoo, J, Wu, M, Yin, Y, Herzik, M.A.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a mitochondrial calcium uniporter.
Science, 361, 2018
|
|
6D73
 
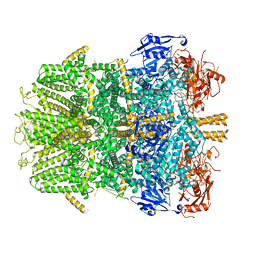 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel, subfamily M | | Authors: | Yin, Y, Wu, M, Borschel, W.F, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
7RQY
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an open state, class B | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQW
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an open state, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQV
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an intermediate-closed state, class II | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQU
 
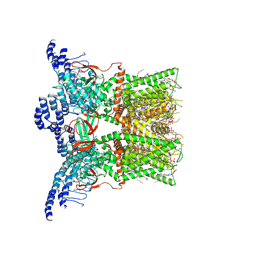 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in a closed state, class I | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, SODIUM ION, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQX
 
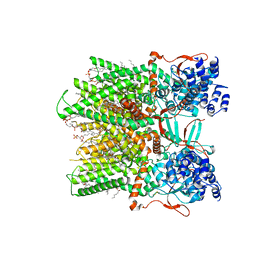 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an intermediate-open state, class A | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQZ
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 48 degrees Celsius, in an open state, class alpha | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
8TZ9
 
 | |
8TZ2
 
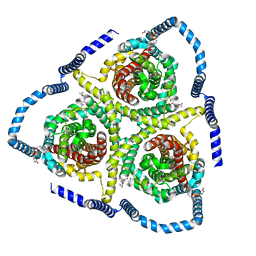 | |
8TZD
 
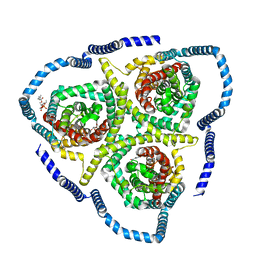 | |
8TZ6
 
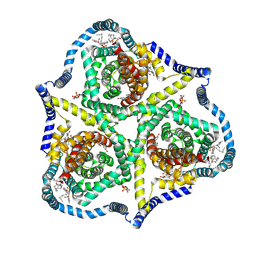 | |
8TZK
 
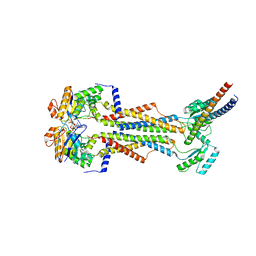 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, shortened | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZJ
 
 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZL
 
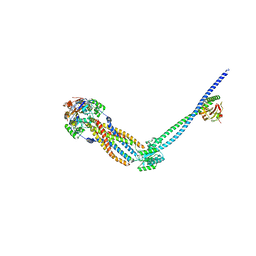 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, full-length | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZ3
 
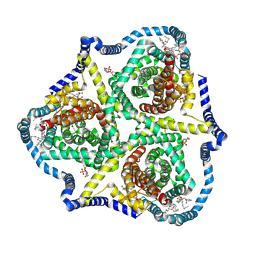 | |
8TZ7
 
 | |
8TZ5
 
 | |
