5DKK
 
 | |
5DKL
 
 | |
8C05
 
 | | LOV-activated diguanylate cyclase, dark-state structure | | Descriptor: | DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Vide, U, Winkler, A. | | Deposit date: | 2022-12-15 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases.
Sci Adv, 9, 2023
|
|
7OO9
 
 | |
5LUV
 
 | |
4WUJ
 
 | | Structural Biochemistry of a Fungal LOV Domain Photoreceptor Reveals an Evolutionarily Conserved Pathway Integrating Blue-Light and Oxidative Stress | | Descriptor: | FLAVIN MONONUCLEOTIDE, Glycoside hydrolase family 15, cellulose signaling associated protein envoy, ... | | Authors: | Hopkins, H.C, Lokhandwala, J, Zoltowski, B.D. | | Deposit date: | 2014-10-31 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Biochemistry of a Fungal LOV Domain Photoreceptor Reveals an Evolutionarily Conserved Pathway Integrating Light and Oxidative Stress.
Structure, 23, 2015
|
|
5J4E
 
 | |
5J3W
 
 | |
3T50
 
 | |
5A8B
 
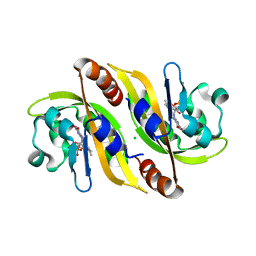 | | Structure of a parallel dimer of the aureochrome 1a LOV domain from Phaeodactylum tricornutum | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Banerjee, A, Herman, E, Kottke, T, Essen, L.O. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Structure of a Native-Like Aureochrome 1A Lov Domain Dimer from Phaeodactylum Tricornutum.
Structure, 24, 2016
|
|
3SW1
 
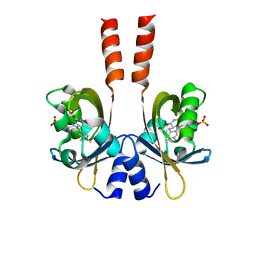 | | Structure of a full-length bacterial LOV protein | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sensory box protein | | Authors: | Granzin, J, Batra-Safferling, R, Jaeger, K.-E, Drepper, T, Krauss, U. | | Deposit date: | 2011-07-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida.
J.Mol.Biol., 417, 2012
|
|
5SVV
 
 | |
5SVU
 
 | |
5SVG
 
 | |
7R5N
 
 | |
6GBV
 
 | |
6GAY
 
 | |
6GB3
 
 | |
6GBA
 
 | |
2YOM
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
4KUO
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
3ULF
 
 | |
3UE6
 
 | |
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
