1KHU
 
 | |
3KMP
 
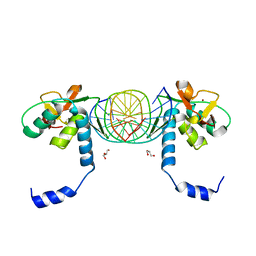 | | Crystal Structure of SMAD1-MH1/DNA complex | | Descriptor: | 5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3', 5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3', GLYCEROL, ... | | Authors: | Baburajendran, N, Palasingam, P, Narasimhan, K, Jauch, R, Kolatkar, P.R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors.
Nucleic Acids Res., 38, 2010
|
|
3Q4A
 
 | |
3Q47
 
 | |
3Q49
 
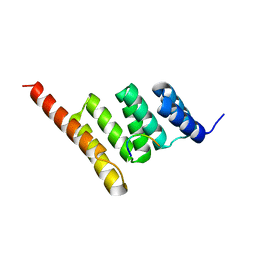 | |
5ZOK
 
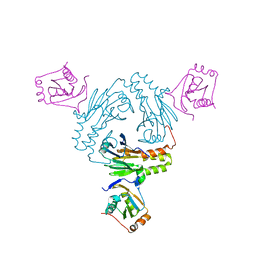 | | Crystal structure of human SMAD1-MAN1 complex. | | Descriptor: | Inner nuclear membrane protein Man1, Mothers against decapentaplegic homolog 1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for receptor-regulated SMAD recognition by MAN1
Nucleic Acids Res., 46, 2018
|
|
2LAW
 
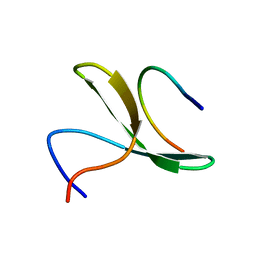 | | Structure of the second WW domain from human YAP in complex with a human Smad1 derived peptide | | Descriptor: | Mothers against decapentaplegic homolog 1, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2LAZ
 
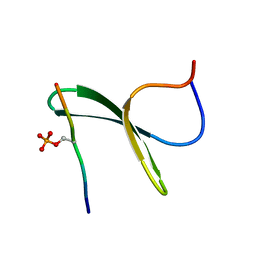 | | Structure of the first WW domain of human Smurf1 in complex with a mono-phosphorylated human Smad1 derived peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF1, Mothers against decapentaplegic homolog 1 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2LB0
 
 | | Structure of the first WW domain of human Smurf1 in complex with a di-phosphorylated human Smad1 derived peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF1, Mothers against decapentaplegic homolog 1 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2LAY
 
 | | Structure of the first WW domain of human YAP in complex with a phosphorylated human Smad1 derived peptide | | Descriptor: | Mothers against decapentaplegic homolog 1, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2LB1
 
 | | Structure of the second domain of human Smurf1 in complex with a human Smad1 derived peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF1, Mothers against decapentaplegic homolog 1 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2LAX
 
 | | Structure of first WW domain of human YAP in complex with a human Smad1 doubly-phosphorilated derived peptide. | | Descriptor: | Mothers against decapentaplegic homolog 1, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
3DIT
 
 | |
5ZOJ
 
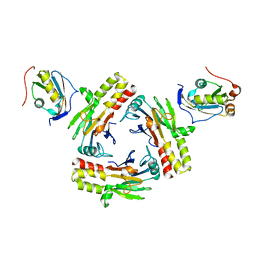 | | Crystal structure of human SMAD2-MAN1 complex | | Descriptor: | Inner nuclear membrane protein Man1, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Ohno, Y, Ito, T, Tanokura, M. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structural basis for receptor-regulated SMAD recognition by MAN1
Nucleic Acids Res., 46, 2018
|
|
3MY0
 
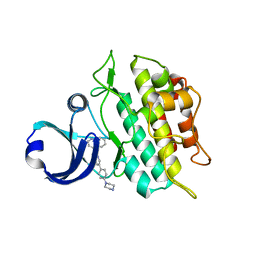 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
4MPL
 
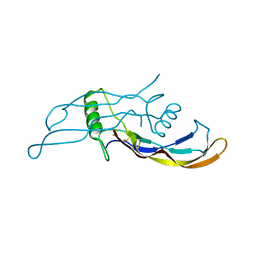 | | Crystal structure of BMP9 at 1.90 Angstrom | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Li, W, Morrell, N.W, Wei, Z. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of Bone Morphogenetic Protein 9 (BMP9) by Redox-dependent Proteolysis.
J.Biol.Chem., 289, 2014
|
|
6ZMN
 
 | |
2CH0
 
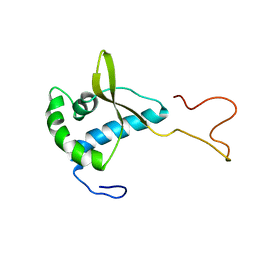 | | Solution structure of the human MAN1 C-terminal domain (residues 655- 775) | | Descriptor: | INNER NUCLEAR MEMBRANE PROTEIN MAN1 | | Authors: | Caputo, S, Couprie, J, Duband-Goulet, I, Lin, F, Braud, S, Gondry, M, Worman, H.J, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain.
J. Biol. Chem., 281, 2006
|
|
2ZPY
 
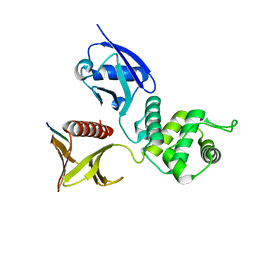 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
6TBZ
 
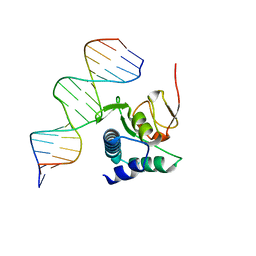 | |
6TCE
 
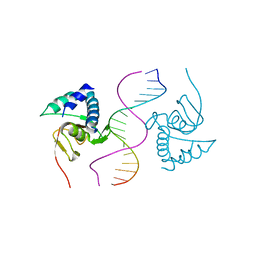 | |
5MEZ
 
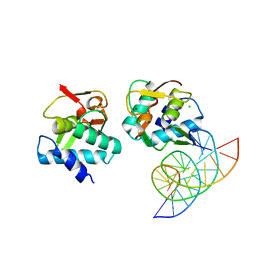 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MEY
 
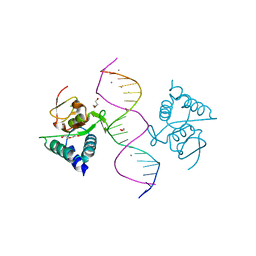 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MF0
 
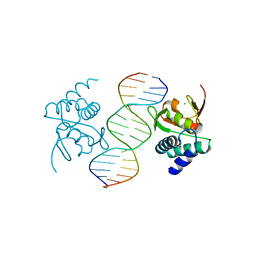 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
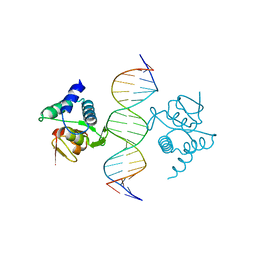 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
