3LIS
 
 | |
3LFP
 
 | |
4JCX
 
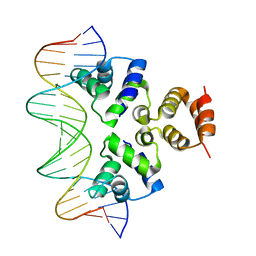 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JCY
 
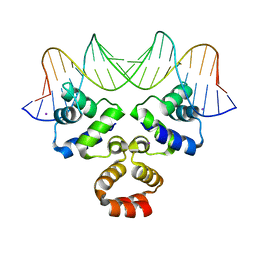 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OR operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*AP*AP*CP*TP*AP*AP*GP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*CP*TP*TP*AP*GP*TP*TP*T)-3'), ... | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JQD
 
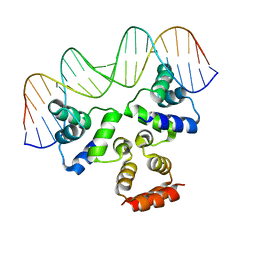 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
