5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
1S6L
 
 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
5C0U
 
 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0T
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
3F0P
 
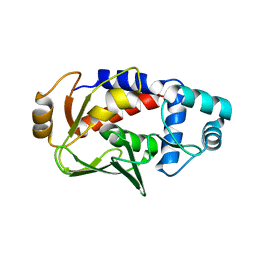 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0O
 
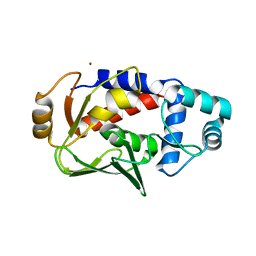 | | Crystal structure of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2H
 
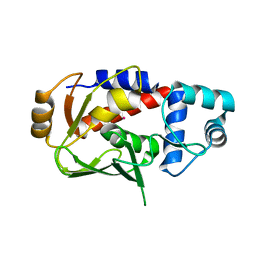 | | Crystal structure of the mercury-bound form of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2G
 
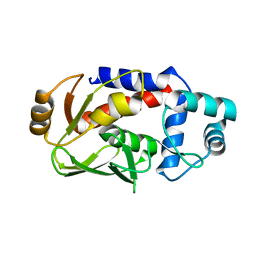 | | Crystal structure of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2F
 
 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3FN8
 
 | |
5U7A
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | Alkylmercury lyase, BROMIDE ION, Dimethyltin dibromide, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U88
 
 | | Crystal structure of a MerB-triimethyllead complex. | | Descriptor: | ACETATE ION, Alkylmercury lyase, Trimethyllead bromide | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-11 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7B
 
 | | Crystal structure of a the tin-bound form of MerB formed from Diethyltin. | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U79
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U83
 
 | | Crystal structure of a MerB-trimethytin complex. | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7C
 
 | | Crystal structure of the lead-bound form of MerB formed from diethyllead. | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U82
 
 | | Crystal structure of a MerB-triethyltin complex | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-01-11 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
