4UXA
 
 | | Improved variant of (R)-selective manganese-dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Pavkov-Keller, T, Wiedner, R, Kothbauer, B, Gruber-Khadjawi, M, Schwab, H, Steiner, K, Gruber, K. | | Deposit date: | 2014-08-21 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the Properties of Bacterial R-Selective Hydroxynitrile Lyases for Industrial Applications
Chemcatchem, 2015
|
|
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | Descriptor: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | Authors: | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
5BPX
 
 | | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP. | | Descriptor: | 2,4'-dihydroxyacetophenone dioxygenase, ACETATE ION, FE (III) ION, ... | | Authors: | Guo, J, Erskine, P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Extension of resolution and oligomerization-state studies of 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7ZVY
 
 | |
7ZVM
 
 | |
2GU9
 
 | |
2H0V
 
 | |
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
2I45
 
 | |
5J4F
 
 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
5J4G
 
 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
5J7M
 
 | | Crystal structure of Cupin 2 conserved barrel domain protein from Kribbella flavida DSM 17836 | | Descriptor: | ACETATE ION, Cupin 2 conserved barrel domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Cuff, M, Chhor, G, Endres, M, Joachimiak, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Cupin 2 conserved barrel domain protein from Kribbella flavida DSM 17836
To Be Published
|
|
6L2F
 
 | | Crystal structure of a cupin protein (tm1459, H54AH58A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, COPPER (II) ION, ... | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | Descriptor: | COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4P9G
 
 | | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. | | Descriptor: | 2,4'-dihydroxyacetophenone dioxygenase, CARBONATE ION, FE (III) ION, ... | | Authors: | Keegan, R, Lebedev, A, Erskine, P, Guo, J, Wood, S.P, Hopper, D.J, Cooper, J.B. | | Deposit date: | 2014-04-03 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
1O4T
 
 | |
6A55
 
 | | Crystal structure of DddK mutant Y122A | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6A54
 
 | | Crystal structure of DddK mutant Y64A | | Descriptor: | MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6A53
 
 | | Crystal structure of DddK | | Descriptor: | MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
1SQ4
 
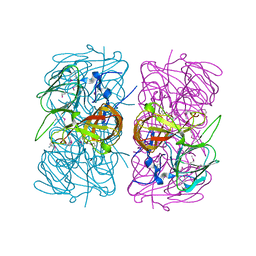 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
8HFB
 
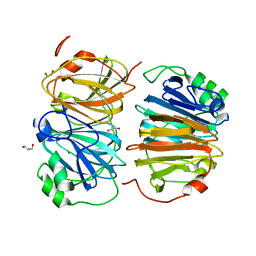 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Eom, H, Song, W.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
3BU7
 
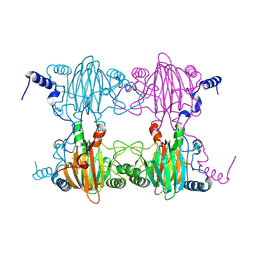 | | Crystal Structure and Biochemical Characterization of GDOsp, a Gentisate 1,2-Dioxygenase from Silicibacter Pomeroyi | | Descriptor: | FE (II) ION, Gentisate 1,2-dioxygenase | | Authors: | Chen, J, Wang, M.Z, Zhu, G.Y, Zhang, X.C, Rao, Z.H. | | Deposit date: | 2008-01-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and mutagenic analysis of GDOsp, a gentisate 1,2-dioxygenase from Silicibacter pomeroyi.
Protein Sci., 17, 2008
|
|
