6HZR
 
 | | Apo structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HR4
 
 | |
7AU1
 
 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 12) | | Descriptor: | 2-(6-(((R)-2-amino-2-oxo-1-phenylethyl)carbamoyl)-1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acetic acid, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7ATW
 
 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 3) | | Descriptor: | 1-Hydroxy-1,3-dihydro-2,1-benzoxaborole-6-carboxylic acid, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | Descriptor: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
7ATM
 
 | | Structure of P. aeruginosa PBP3 in complex with a phenyl boronic acid (Compound 1) | | Descriptor: | (3-(1H-tetrazol-5-yl)phenyl)boronic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-10-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7ATO
 
 | | Structure of P. aeruginosa PBP3 in complex with an aryl boronic acid (Compound 2) | | Descriptor: | (5-methyl-1H-indazol-6-yl)boronic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-10-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
6R3X
 
 | |
6I1E
 
 | | Crystal structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 in complex with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
3PBT
 
 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5UY7
 
 | |
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
3PBQ
 
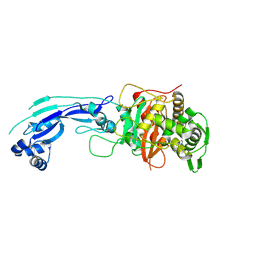 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6R42
 
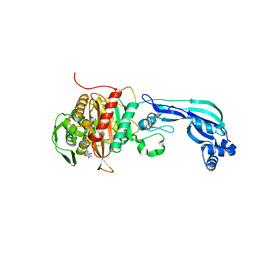 | |
7ONK
 
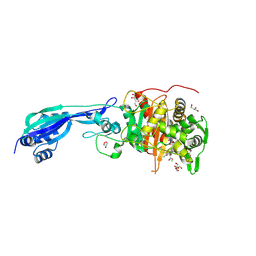 | | Crystal structure of PBP3 from P. aeruginosa in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
4WEK
 
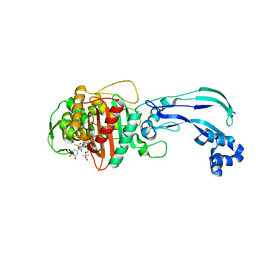 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
3PBO
 
 | | Crystal structure of PBP3 complexed with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
7ONY
 
 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7AU8
 
 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 13) | | Descriptor: | 2-(1-hydroxy-6-((2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl)carbamoyl)-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acetic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7ATX
 
 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 4) | | Descriptor: | 4-(1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborole-6-carbonyl)-1,3,3-trimethylpiperazin-2-one, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
5TRO
 
 | | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.
To Be Published
|
|
5TT1
 
 | |
1VQQ
 
 | |
6BSQ
 
 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
