1BJC
 
 | | SOLUTION NMR STRUCTURE OF AMYLOID BETA[F16], RESIDUES 1-28, 15 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Poulsen, S.-A, Watson, A.A, Craik, D.J. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structures in aqueous SDS micelles of two amyloid beta peptides of A beta(1-28) mutated at the alpha-secretase cleavage site (K16E, K16F)
J.Struct.Biol., 130, 2000
|
|
5OQV
 
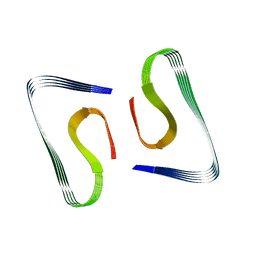 | | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | | Descriptor: | Amyloid beta A4 protein | | Authors: | Gremer, L, Schoelzel, D, Schenk, C, Reinartz, E, Labahn, J, Ravelli, R, Tusche, M, Lopez-Iglesias, C, Hoyer, W, Heise, H, Willbold, D, Schroeder, G.F. | | Deposit date: | 2017-08-14 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Fibril structure of amyloid-beta (1-42) by cryo-electron microscopy.
Science, 358, 2017
|
|
3IFN
 
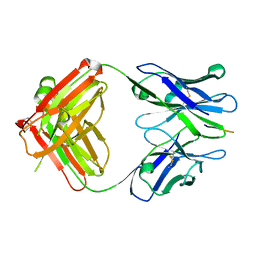 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-40:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
2OTK
 
 | |
2BEG
 
 | | 3D Structure of Alzheimer's Abeta(1-42) fibrils | | Descriptor: | Amyloid beta A4 protein | | Authors: | Luhrs, T, Ritter, C, Adrian, M, Riek-Loher, D, Bohrmann, B, Dobeli, H, Schubert, D, Riek, R. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-22 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | 3D structure of Alzheimer's amyloid-{beta}(1-42) fibrils.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5HOX
 
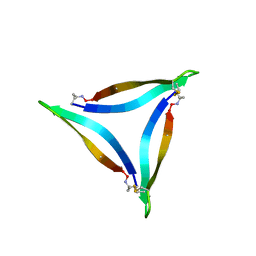 | | X-ray crystallographic structure of an A-beta 17_36 beta-hairpin. Synchrotron data set. (LVFFAEDCGSNKCAII(SAR)LMV). | | Descriptor: | Amyloid beta A4 protein, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Kreutzer, A.G, Nowick, J.S, Spencer, R.K. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an A beta 17-36 beta-Hairpin.
J.Am.Chem.Soc., 138, 2016
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
1Z0Q
 
 | | Aqueous Solution Structure of the Alzheimer's Disease Abeta Peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Tomaselli, S, Esposito, V, Vangone, P, van Nuland, N.A, Bonvin, A.M, Guerrini, R, Tancredi, T, Temussi, P.A, Picone, D. | | Deposit date: | 2005-03-02 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The alpha-to-beta Conformational Transition of Alzheimer's Abeta-(1-42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of beta Conformation Seeding
Chembiochem, 7, 2006
|
|
7RTZ
 
 | |
1IYT
 
 | | Solution structure of the Alzheimer's disease amyloid beta-peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Crescenzi, O, Tomaselli, S, Guerrini, R, Salvadori, S, D'Ursi, A.M, Temussi, P.A, Picone, D. | | Deposit date: | 2002-09-06 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain.
EUR.J.BIOCHEM., 269, 2002
|
|
7Q4M
 
 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q4B
 
 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
5MYK
 
 | |
6W0O
 
 | |
5MYX
 
 | |
6WXM
 
 | |
7U4P
 
 | |
7Y8Q
 
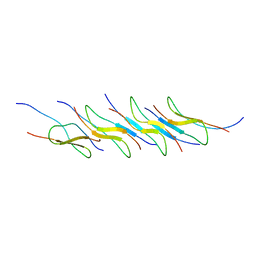 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
1NMJ
 
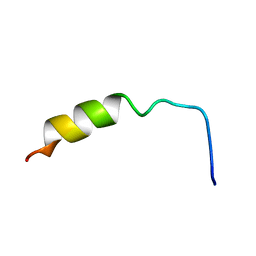 | |
7B3K
 
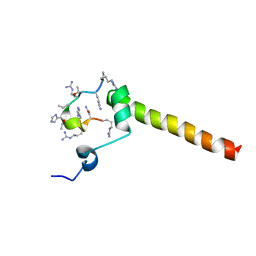 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7B3J
 
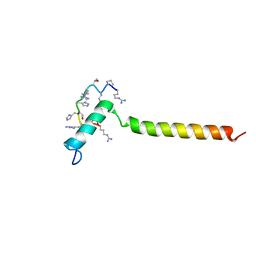 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
5HOW
 
 | |
6NB9
 
 | | Amyloid-Beta (20-34) with L-isoaspartate 23 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Boyer, D.R, Zee, C.T, Richards, L.S, Cascio, D, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-08-07 | | Last modified: | 2022-09-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
5HOY
 
 | | X-ray crystallographic structure of an A-beta 17_36 beta-hairpin. X-ray diffractometer data set. (LVFFAEDCGSNKCAII(SAR)LMV). | | Descriptor: | Amyloid beta A4 protein, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an A beta 17-36 beta-Hairpin.
J.Am.Chem.Soc., 138, 2016
|
|
7F29
 
 | |
