2YYY
 
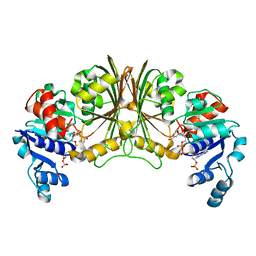 | | Crystal structure of Glyceraldehyde-3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malay, A.D, Bessho, Y, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from the archaeal hyperthermophile Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1A7K
 
 | | GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN A MONOCLINIC CRYSTAL FORM | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Hol, W.G.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase in a new crystal form confirms the putative physiological active site structure.
J.Mol.Biol., 278, 1998
|
|
5ZA0
 
 | |
6QUN
 
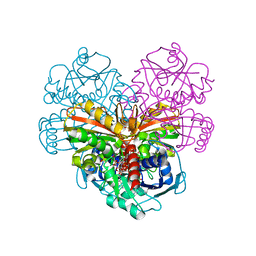 | | Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QUQ
 
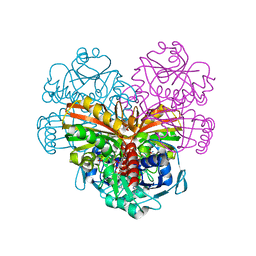 | | Crystal structure of glutathionylated glycolytic glyceraldehyde-3- phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1) | | Descriptor: | GLUTATHIONE, Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WNI
 
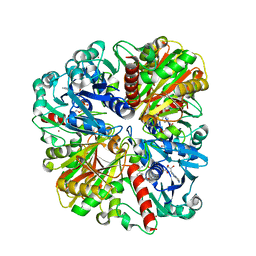 | |
4WNC
 
 | | Crystal structure of human wild-type GAPDH at 1.99 angstroms resolution | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Garcin, E.D. | | Deposit date: | 2014-10-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Dimer Interface Mutation in Glyceraldehyde-3-Phosphate Dehydrogenase Regulates Its Binding to AU-rich RNA.
J.Biol.Chem., 290, 2015
|
|
1B7G
 
 | | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE | | Descriptor: | PROTEIN (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Isupov, M.N, Littlechild, J.A. | | Deposit date: | 1999-01-22 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J.Mol.Biol., 291, 1999
|
|
6ADE
 
 | | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dilawari, R, Singh, P.K, Raje, M, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution
To Be Published
|
|
7JH0
 
 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Schistosoma mansoni | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Boreiko, S, Silva, M, Iulek, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure determination and analyses of the GAPDH from the parasite Schistosoma mansoni, the first one from a platyhelminth.
Biochimie, 184, 2021
|
|
7U4S
 
 | | Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Miranda, R.R, Silva, M, Iulek, J. | | Deposit date: | 2022-02-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Expression, purification, crystallization and structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans, main causative agent of candidiasis
Chem. Data Coll., 39, 2022
|
|
7JWK
 
 | |
7U5M
 
 | | Cryo-EM Structure of GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgan, C.E, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
5LD5
 
 | | Crystal structure of a bacterial dehydrogenase at 2.19 Angstroms resolution | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Querol-Garcia, J, Fernandez, F.J, Gomez, S, Fulla, D, Juanhuix, J, Vega, M.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.1906 Å) | | Cite: | Crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from the Gram-Positive Bacterial Pathogen A. vaginae, an Immunoevasive Factor that Interacts with the Human C5a Anaphylatoxin.
Front Microbiol, 8, 2017
|
|
6NLX
 
 | |
6OK4
 
 | |
5M6D
 
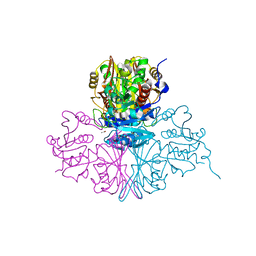 | | Streptococcus pneumoniae Glyceraldehyde-3-Phosphate Dehydrogenase (SpGAPDH) crystal structure | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gaboriaud, C, Moreau, C.P, Di Guilmi, A.M. | | Deposit date: | 2016-10-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering Key Residues Involved in the Virulence-promoting Interactions between Streptococcus pneumoniae and Human Plasminogen.
J. Biol. Chem., 292, 2017
|
|
4GPD
 
 | |
4BOY
 
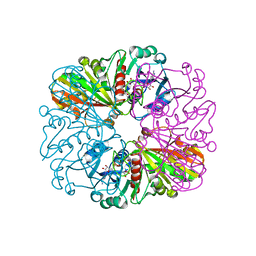 | |
8BR4
 
 | |
2PKR
 
 | | Crystal structure of (A+CTE)4 chimeric form of photosyntetic glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase Aor, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PKQ
 
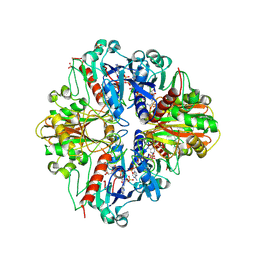 | | Crystal structure of the photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, Glyceraldehyde-3-phosphate dehydrogenase B, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6PX2
 
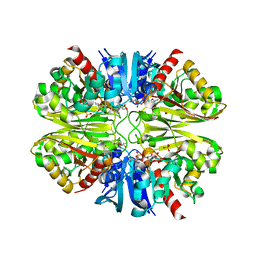 | | Acropora millepora GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Brandt, G.S, Fields, P.A. | | Deposit date: | 2019-07-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora.
Rsc Adv, 11, 2021
|
|
5O0V
 
 | |
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
