5GSL
 
 | | Glycoside hydrolase A | | Descriptor: | 778aa long hypothetical beta-galactosidase, PHOSPHATE ION | | Authors: | Watanabe, M, Kamachi, S, Mine, S. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycoside hydrolase A
To Be Published
|
|
5GSM
 
 | | Glycoside hydrolase B with product | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Watanabe, M, Kamachi, S, Mine, S. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Glycoside hydrolase B with product
To Be Published
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | Descriptor: | B-agarase | | Authors: | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
3U7V
 
 | |
8FA4
 
 | |
5JAW
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,4S,5R,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2016-04-12 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards broad spectrum activity-based glycosidase probes: synthesis and evaluation of deoxygenated cyclophellitol aziridines.
Chem. Commun. (Camb.), 53, 2017
|
|
4OIF
 
 | | 3D structure of Gan42B, a GH42 beta-galactosidase from G. | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Feinberg, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of GanB, a GH42 intracellular beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
6JOW
 
 | |
1KWK
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
1KWG
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
6LVW
 
 | |
5DFA
 
 | | 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Lansky, S, Feinberg, H, Govada, L, Chayen, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2015-08-26 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function relationships in Gan42B, an intracellular GH42 beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5E9A
 
 | |
8IBT
 
 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E318S mutant in complex with lacto-N-tetraose | | Descriptor: | Beta-galactosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBS
 
 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBR
 
 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis in complex with glycerol | | Descriptor: | Beta-galactosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8PEJ
 
 | | CjGH35 with a Galactosidase Activity-Based Probe | | Descriptor: | (1~{S},2~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-06-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Org.Biomol.Chem., 21, 2023
|
|
4BQ4
 
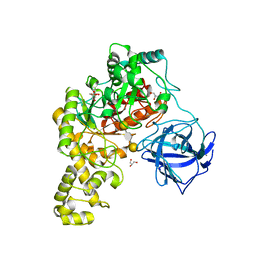 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, B-AGARASE, CALCIUM ION, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4BQ5
 
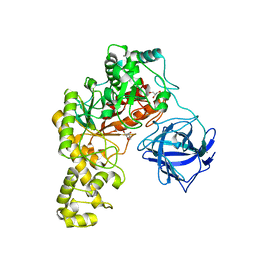 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4BQ2
 
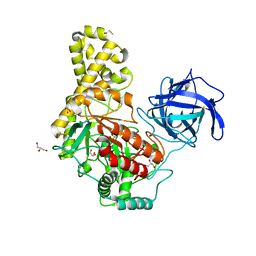 | |
4BQ3
 
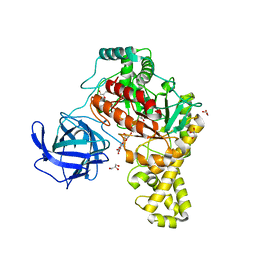 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4D1J
 
 | | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Larsbrink, J, Thompson, A.J, Lundqvist, M, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complex Gene Locus Enables Xyloglucan Utilization in the Model Saprophyte Cellvibrio Japonicus.
Mol.Microbiol., 94, 2014
|
|
4D1I
 
 | | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Thompson, A.J, Lundqvist, M, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complex Gene Locus Enables Xyloglucan Utilization in the Model Saprophyte Cellvibrio Japonicus.
Mol.Microbiol., 94, 2014
|
|
6TBK
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{R},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
