6DVV
 
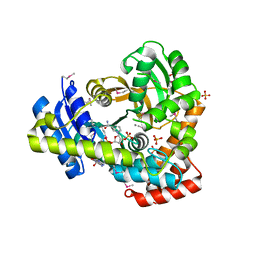 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
3U95
 
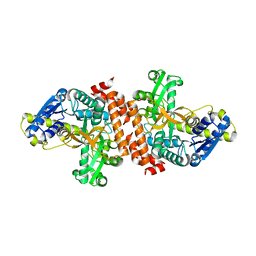 | | Crystal structure of a putative alpha-glucosidase from Thermotoga neapolitana | | Descriptor: | Glycoside hydrolase, family 4, MANGANESE (II) ION | | Authors: | Ha, N.C, Jun, S.Y, Yun, B.Y, Yoon, B.Y, Piao, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure and thermostability of a putative alpha-glucosidase from Thermotoga neapolitana
Biochem.Biophys.Res.Commun., 416, 2011
|
|
6KCX
 
 | |
3FEF
 
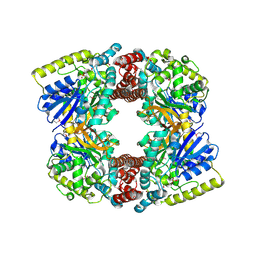 | | Crystal structure of putative glucosidase lplD from bacillus subtilis | | Descriptor: | MAGNESIUM ION, Putative glucosidase lplD, ALPHA-GALACTURONIDASE, ... | | Authors: | Ramagopal, U.A, Rajashankar, K.R, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glucosidase lplD from bacillus subtilis.
To be published
|
|
5C3M
 
 | | Crystal structure of Gan4C, a GH4 6-phospho-glucosidase from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Putative 6-phospho-beta-glucosidase | | Authors: | Cohen, T, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.059 Å) | | Cite: | Crystal structure of Gan4C, a GH4 6-phospho-glucosidase from Geobacillus stearothermophilus
To Be Published
|
|
6DUX
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 6-phospho-alpha-glucosidase, ACETATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-04 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
1OBB
 
 | | alpha-glucosidase A, AglA, from Thermotoga maritima in complex with maltose and NAD+ | | Descriptor: | ALPHA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lodge, J.A, Maier, T, Liebl, W, Hoffmann, V, Strater, N. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Thermotoga Maritima Alpha-Glucosidase Agla Defines a New Clan of Nad+-Dependent Glycosidases
J.Biol.Chem., 278, 2003
|
|
1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
1U8X
 
 | | CRYSTAL STRUCTURE OF GLVA FROM BACILLUS SUBTILIS, A METAL-REQUIRING, NAD-DEPENDENT 6-PHOSPHO-ALPHA-GLUCOSIDASE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, Maltose-6'-phosphate glucosidase, ... | | Authors: | Rajan, S.S, Yang, X, Collart, F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel Catalytic Mechanism of Glycoside Hydrolysis Based on the Structure of an NAD(+)/Mn(2+)-Dependent Phospho-alpha-Glucosidase from Bacillus subtilis.
STRUCTURE, 12, 2004
|
|
1UP7
 
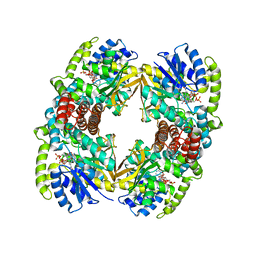 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1UP4
 
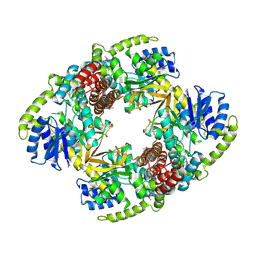 | |
1UP6
 
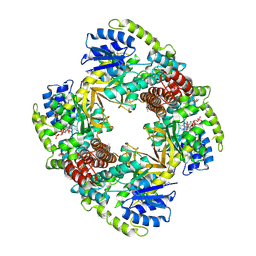 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.55 Angstrom resolution in the tetragonal form with manganese, NAD+ and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, MANGANESE (II) ION, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An Unusual Mechanism of Glycoside Hydrolysis Involving Redox and Elimination Steps by a Family 4 Beta-Glycosidase from Thermotoga Maritima.
J.Am.Chem.Soc., 126, 2004
|
|
1VJT
 
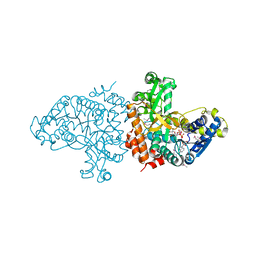 | |
7BR4
 
 | |
7BRF
 
 | |
7CTD
 
 | |
7CTL
 
 | |
7CTM
 
 | |
