1E0E
 
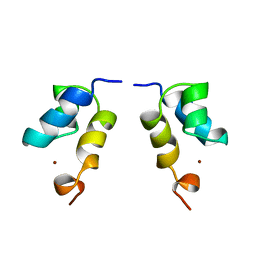 | | N-terminal zinc-binding HHCC domain of HIV-2 integrase | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 INTEGRASE, ZINC ION | | Authors: | Eijkelenboom, A.P.A.M, Van Den ent, F.M.I, Plasterk, R.H.A, Kaptein, R, Boelens, R. | | Deposit date: | 2000-03-25 | | Release date: | 2001-03-19 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the Dimeric N-Terminal Hhcc Domain of HIV-2 Integrase
J.Biomol.NMR, 18, 2000
|
|
1K6Y
 
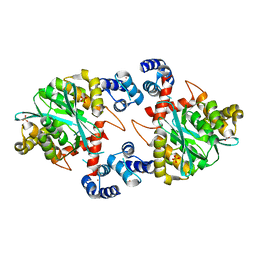 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
1WJA
 
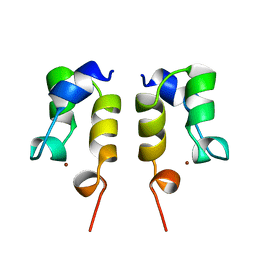 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJB
 
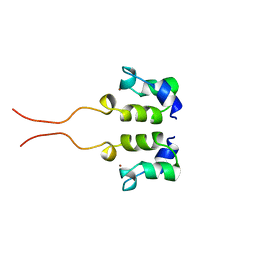 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJC
 
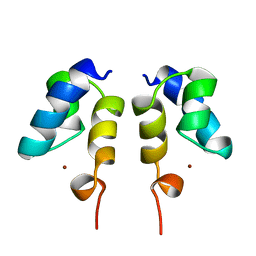 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
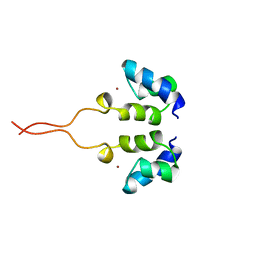 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJE
 
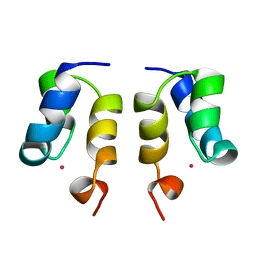 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1WJF
 
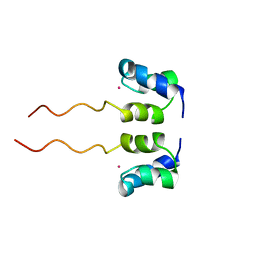 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
3F9K
 
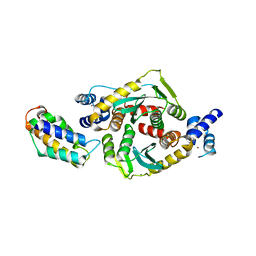 | | Two domain fragment of HIV-2 integrase in complex with LEDGF IBD | | Descriptor: | Integrase, MAGNESIUM ION, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75
Plos Pathog., 5, 2009
|
|
3HPG
 
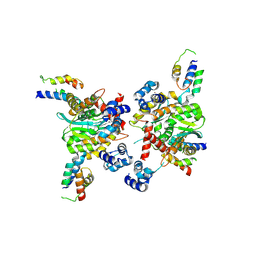 | |
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
3JCA
 
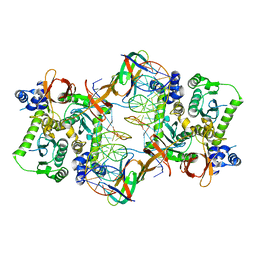 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
4FW2
 
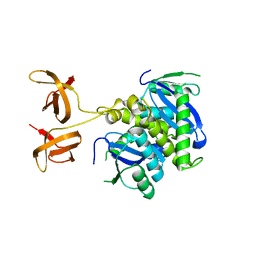 | |
5CZ2
 
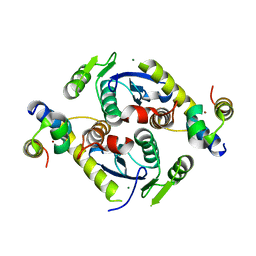 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
5EJK
 
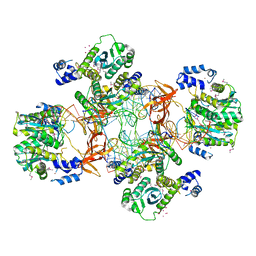 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
5HOT
 
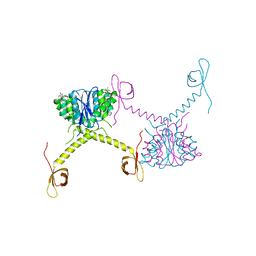 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | Authors: | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5M0R
 
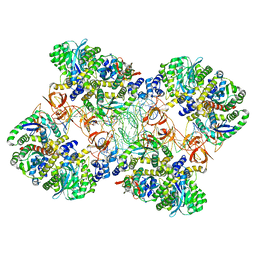 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
5U1C
 
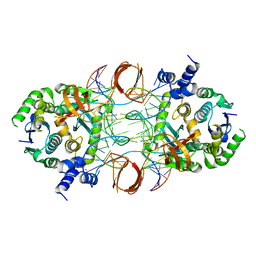 | |
6PUT
 
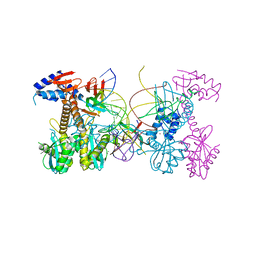 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | | Descriptor: | CALCIUM ION, Chimeric Sso7d and HIV-1 integrase, ZINC ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUW
 
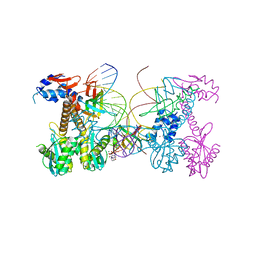 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | Descriptor: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
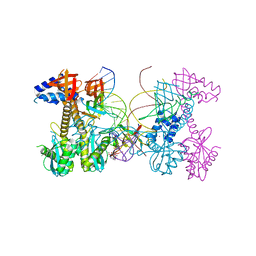 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUZ
 
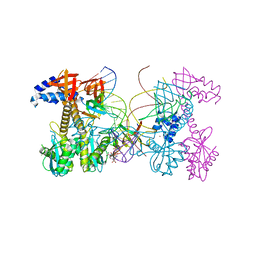 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
