1C3G
 
 | | S. CEREVISIAE HEAT SHOCK PROTEIN 40 SIS1 | | Descriptor: | HEAT SHOCK PROTEIN 40 | | Authors: | Sha, B, Lee, S, Cyr, D. | | Deposit date: | 1999-07-27 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1.
Structure, 8, 2000
|
|
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
1NLT
 
 | | The crystal structure of Hsp40 Ydj1 | | Descriptor: | Mitochondrial protein import protein MAS5, Seven residue peptide, ZINC ION | | Authors: | Li, J, Sha, B. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.
Structure, 11, 2003
|
|
1XAO
 
 | | Hsp40-Ydj1 dimerization domain | | Descriptor: | Mitochondrial protein import protein MAS5 | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2004-08-26 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40
J.Mol.Biol., 346, 2005
|
|
2CTT
 
 | | Solution structure of zinc finger domain from human DnaJ subfamily A menber 3 | | Descriptor: | DnaJ homolog subfamily A member 3, ZINC ION | | Authors: | Kobayashi, N, Tochio, N, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain from human DnaJ subfamily A menber 3
To be Published
|
|
2B26
 
 | | The crystal structure of the protein complex of yeast Hsp40 Sis1 and Hsp70 Ssa1 | | Descriptor: | Heat shock 70 kDa protein cognate 2, SIS1 protein | | Authors: | Li, J, Wu, Y, Qian, X, Sha, B. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex.
Biochem.J., 398, 2006
|
|
2Q2G
 
 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
2QLD
 
 | | human Hsp40 Hdj1 | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Hu, J, Wu, Y, Li, J, Fu, Z, Sha, B. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1.
Bmc Struct.Biol., 8, 2008
|
|
3I38
 
 | | Structure of a putative chaperone protein dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Putative Chaperone Protein Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578
To be Published
|
|
3LZ8
 
 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
3AGX
 
 | |
3AGY
 
 | |
3AGZ
 
 | |
4J80
 
 | | Thermus thermophilus DnaJ | | Descriptor: | Chaperone protein DnaJ 2 | | Authors: | Barends, T.R.M, Brosi, R.W, Steinmetz, A, Scherer, A, Hartmann, E, Eschenbach, J, Lorenz, T, Seidel, R, Shoeman, R, Zimmermann, S, Bittl, R, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-31 | | Last modified: | 2013-10-09 | | Method: | EPR (2.9 Å), X-RAY DIFFRACTION | | Cite: | Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6JZB
 
 | |
6PPT
 
 | |
6PQ2
 
 | |
6PQE
 
 | |
6PQM
 
 | |
6PRI
 
 | |
6PRP
 
 | |
6PRQ
 
 | |
6PRJ
 
 | |
6PSI
 
 | |
7JTK
 
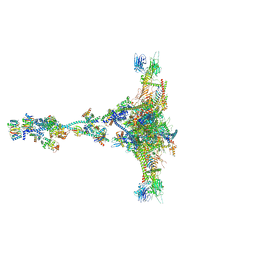 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
