8YDO
 
 | | Crystal structure of dKeima570 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dKeima570
To Be Published
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8T6L
 
 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
8SOK
 
 | |
8SOJ
 
 | |
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | Descriptor: | CHLORIDE ION, SULFATE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 2024
|
|
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 2024
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 2024
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8OVP
 
 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
8OVO
 
 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVN
 
 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OTS
 
 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSI
 
 | | Genetically encoded green ratiometric calcium indicator FNCaMP in calcium-bound state | | Descriptor: | CALCIUM ION, mNeonGreen,Calmodulin,Protein kinase domain-containing protein, {(4Z)-2-(aminomethyl)-4-[(4-hydroxyphenyl)methylidene]-5-oxo-4,5-dihydro-1H-imidazol-1-yl}acetic acid | | Authors: | Varfolomeeva, L.A, Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FNCaMP, ratiometric green calcium indicator based on mNeonGreen protein.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
8J3J
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S, Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2L
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (N137A, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2K
 
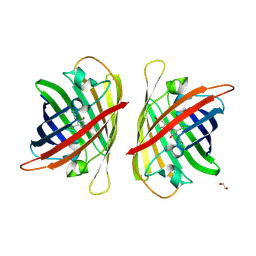 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutation (N137A, Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(N137A, Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2J
 
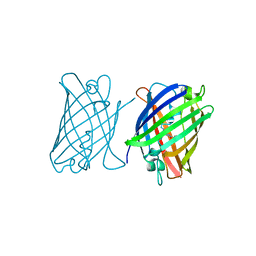 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, StayGold(Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2I
 
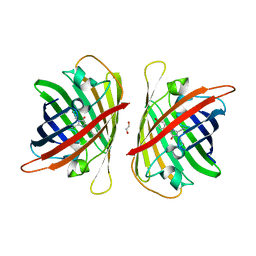 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2H
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (N137A) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | GLYCEROL, SODIUM ION, StayGold(N137A) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J1E
 
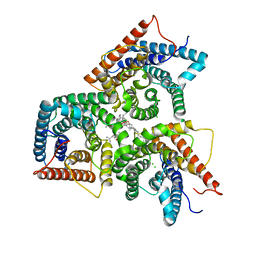 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IM1
 
 | | mCherry-LaM1 complex | | Descriptor: | LaM1, MCherry fluorescent protein, SULFATE ION | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
