7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6IDW
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2018-09-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XYH
 
 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XCY
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5JX5
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo- and endo- activity in a complex with cellobiose
Acta Crystallogr.,Sect.D, 2019
|
|
4AVO
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose | | Descriptor: | ACETATE ION, BETA-1,4-EXOCELLULASE, CALCIUM ION, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4AVN
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D226A- S232A cocrystallized with cellobiose | | Descriptor: | CALCIUM ION, CELLOBIOHYDROLASE. GLYCOSYL HYDROLASE FAMILY 6, beta-D-glucopyranose, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4I5U
 
 | | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chimeric cel6A, ... | | Authors: | Arnold, F.H, Wu, I. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Biotechnol.Bioeng., 110, 2013
|
|
4I5R
 
 | | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chimeric cel6A, ... | | Authors: | Arnold, F.H, Wu, I. | | Deposit date: | 2012-11-28 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Biotechnol.Bioeng., 110, 2013
|
|
4AX6
 
 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AX7
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AU0
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4B4F
 
 | | Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose | | Descriptor: | BETA-1,4-EXOCELLULASE, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4B4H
 
 | | Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain | | Descriptor: | BETA-1,4-EXOCELLULASE | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4A05
 
 | | Structure of the catalytic core domain of the cellobiohydrolase, Cel6A, from Chaetomium thermophilum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE FAMILY 6, LITHIUM ION, ... | | Authors: | Thompson, A.J, Wilson, K.S, Davies, G.J. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Catalytic Core Module of the Chaetomium Thermophilum Family Gh6 Cellobiohydrolase Cel6A.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VOF
 
 | | Cellobiohydrolase mutant, CcCel6C D102A, in the closed form | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOH
 
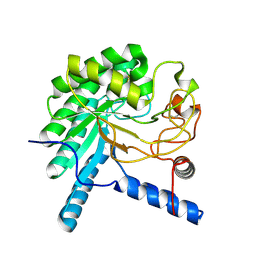 | | CcCel6A catalytic domain complexed with cellobiose | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOJ
 
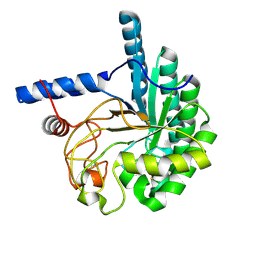 | | CcCel6A catalytic domain mutant D164A | | Descriptor: | Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOG
 
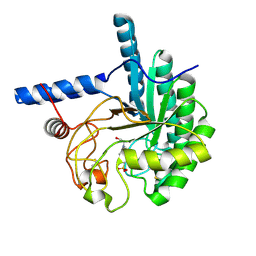 | | Catalytic domain of the cellobiohydrolase, CcCel6A, from Coprinopsis cinerea | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOI
 
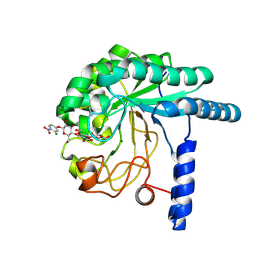 | | CcCel6A catalytic domain complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3RPT
 
 | |
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3A9B
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
