4MSS
 
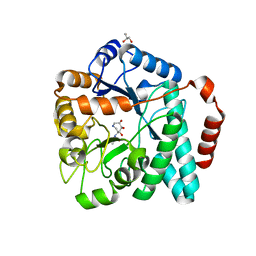 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
2OXN
 
 | |
6DTE
 
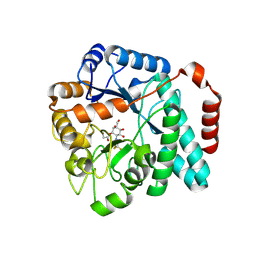 | | GlcNAc-inspired cyclophellitol bound to NagZ | | Descriptor: | 2,2,2-trifluoro-N-[(1R,2R,3R,4R,5R,6R)-2,3,5,6-tetrahydroxy-4-(hydroxymethyl)cyclohexyl]acetamide, Beta-hexosaminidase | | Authors: | Mark, B.L, Winogrodzki, J.L. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | A mechanism-based GlcNAc-inspired cyclophellitol inactivator of the peptidoglycan recycling enzyme NagZ reverses resistance to beta-lactams in Pseudomonas aeruginosa.
Chem. Commun. (Camb.), 54, 2018
|
|
7VI6
 
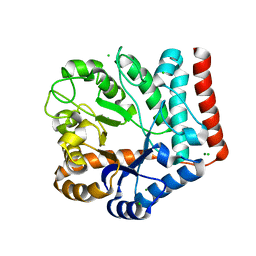 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI7
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
6GFV
 
 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
3GS6
 
 | | Vibrio Cholerea family 3 glycoside hydrolase (NagZ)in complex with N-butyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
3GSM
 
 | | Vibrio cholerae family 3 glycoside hydrolase (NagZ) bound to N-Valeryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-(pentanoylamino)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
1TR9
 
 | |
1Y65
 
 | |
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
5G6T
 
 | | Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-17 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G5U
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-06-05 | | Release date: | 2017-05-17 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G2M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-09 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G3R
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine and L-Ala-1,6-anhydroMurNAc | | Descriptor: | 2-[[(2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoyl]amino]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-30 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G5K
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G1M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, BETA-HEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5UTQ
 
 | |
5UTP
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to N-ethylbutyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, N-[(2Z,3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[(phenylcarbamoyl)oxy]imino}tetrahydro-2H-pyran-3-yl]-2-ethylbutanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5IOB
 
 | | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase-related glycosidases, CHLORIDE ION, ... | | Authors: | Chang, C, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum
To Be Published
|
|
5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5VQE
 
 | | Beta-glucoside phosphorylase BglX bound to 2FGlc | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucoside phosphorylase BglX | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
5VQD
 
 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
