8VEQ
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with azlocillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEP
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 acylated by piperacillin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Piperacillin (Open Form), Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEN
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with cefoperazone | | Descriptor: | (2R,4R)-2-[(1R)-1-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methylidene-1,3-thiazinane-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VBZ
 
 | |
8U55
 
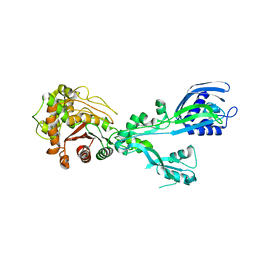 | |
8TJ3
 
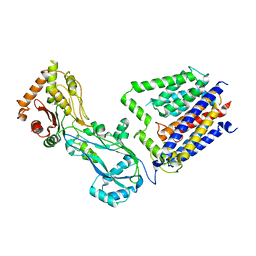 | |
8SQG
 
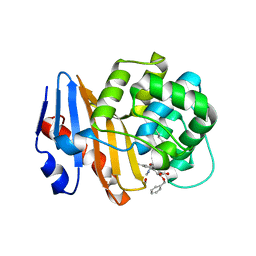 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQF
 
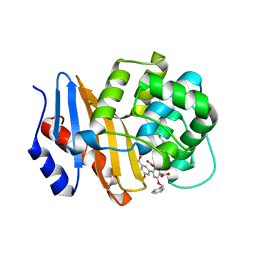 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8RTZ
 
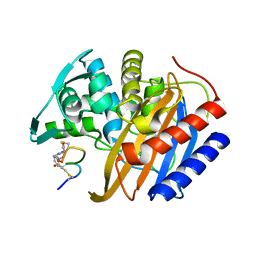 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
8QNZ
 
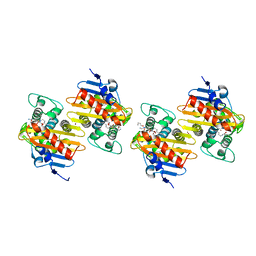 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2023-09-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity.
Acs Cent.Sci., 9, 2023
|
|
8PEC
 
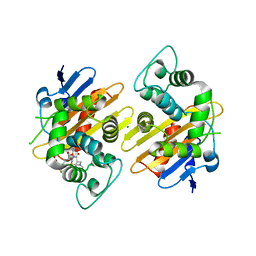 | |
8PEB
 
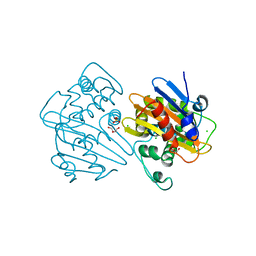 | |
8PEA
 
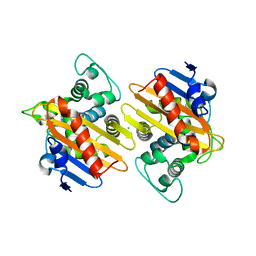 | |
8GPW
 
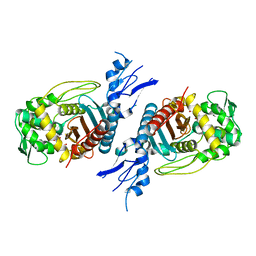 | | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G | | Descriptor: | 1-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2~{S})-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxycyclopropane-1-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Song, D.Q, Li, Y.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G at 2.06 Angstroms resolution
To Be Published
|
|
8FAJ
 
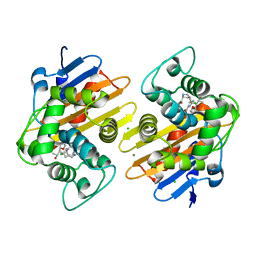 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
8F67
 
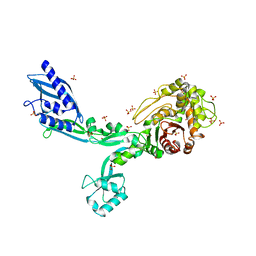 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | Descriptor: | Pbp5, SULFATE ION | | Authors: | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Z
 
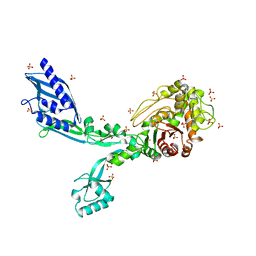 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S422A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Y
 
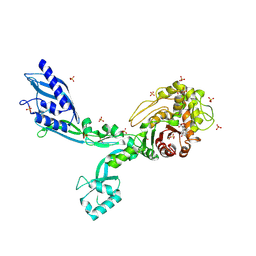 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Poly-Gly variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3X
 
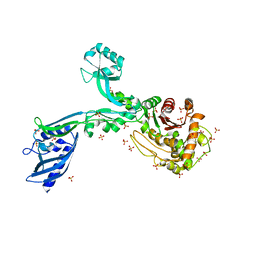 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Poly-Gly variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3W
 
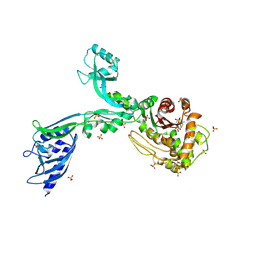 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) PAPAPAP variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3V
 
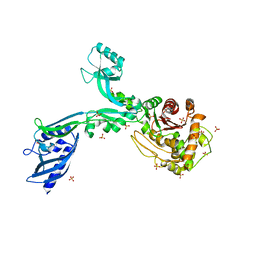 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) PAPAPAP variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3U
 
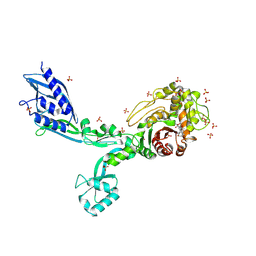 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3T
 
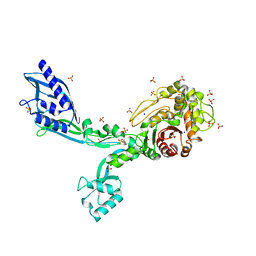 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3S
 
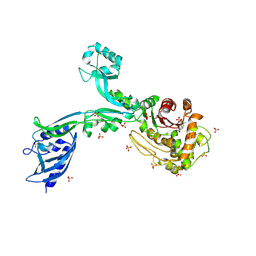 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3R
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
