2J6W
 
 | | R164N mutant of the RUNX1 Runt domain | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Grembecka, J, Zhe, L, Lukasik, S.M, Liu, Y, Bielnicka, I, Bushweller, J.H, Speck, N.A. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mutation in the S-Switch Region of the Runt Domain Alters the Dynamics of an Allosteric Network Responsible for Cbfbeta Regulation.
J.Mol.Biol., 364, 2006
|
|
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
1CO1
 
 | | FOLD OF THE CBFA | | Descriptor: | CORE BINDING FACTOR ALPHA | | Authors: | Berardi, M.J, Bushweller, J.H. | | Deposit date: | 1999-05-31 | | Release date: | 2000-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Ig fold of the core binding factor alpha Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1HJC
 
 | |
1LJM
 
 | | DNA recognition is mediated by conformational transition and by DNA bending | | Descriptor: | CHLORIDE ION, RUNX1 transcription factor | | Authors: | Bartfeld, D, Shimon, L, Couture, G.C, Rabinovich, D, Frolow, F, Levanon, D, Groner, Y, Shakked, Z. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Recognition by the RUNX1 Transcription Factor Is Mediated by an Allosteric Transition in the RUNT Domain and by DNA Bending.
Structure, 10
|
|
1EAQ
 
 | | The RUNX1 Runt domain at 1.25A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EAO
 
 | | THE RUNX1 Runt domain at 1.4A resolution: a structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | BROMIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T.H, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Runx1 Runt Domain at 1.25A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EAN
 
 | | THE RUNX1 Runt domain at 1.70A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-13 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1HJB
 
 | |
1IO4
 
 | |
1E50
 
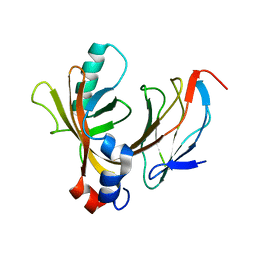 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
1H9D
 
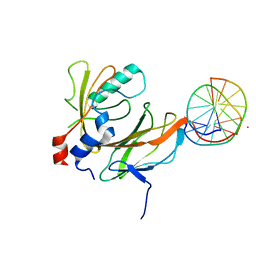 | | Aml1/cbf-beta/dna complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT1, CORE-BINDING FACTOR CBF-BETA, DNA (5'-(*CP*AP*AP*CP*CP*GP*CP*AP*AP*C)-3'), ... | | Authors: | Bravo, J, Warren, A.J. | | Deposit date: | 2001-03-07 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Leukemia-Associated Aml1 (Runx1)-Cbfbeta Complex Functions as a DNA-Induced Molecular Clamp
Nat.Struct.Biol., 8, 2001
|
|
4L0Y
 
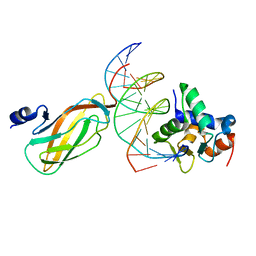 | |
4L18
 
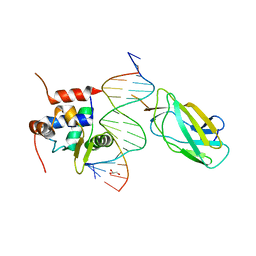 | |
4L0Z
 
 | |
6VGD
 
 | |
6VGG
 
 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2020-01-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
6VGE
 
 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
6VG8
 
 | |
3WTW
 
 | | Crystal structure of the complex comprised of ETS1(K167A), RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3WTY
 
 | | Crystal structure of the complex comprised of ETS1(G333P), RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3WTX
 
 | | Crystal structure of the complex comprised of ETS1(Y329A), RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3WTV
 
 | | Crystal structure of the complex comprised of ETS1(V170G), RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3WTT
 
 | | Crystal structure of the complex comprised of phosphorylated ETS1, RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | 5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3', 5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3', Core-binding factor subunit beta, ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
3WTS
 
 | | Crystal structure of the complex comprised of ETS1, RUNX1, CBFBETA, and the tcralpha gene enhancer DNA | | Descriptor: | 5'-D(*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3', 5'-D(*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3', Core-binding factor subunit beta, ... | | Authors: | Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
