2AYH
 
 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
6VHO
 
 | |
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
5JVV
 
 | | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase | | Descriptor: | beta-1,3-glucosyltransferase | | Authors: | Qin, Z, Yan, Q, Yang, S, Jiang, Z. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Catalytic Mechanism of a Novel Glycoside Hydrolase Family 16 "Elongating" beta-Transglycosylase
J. Biol. Chem., 292, 2017
|
|
5WUT
 
 | |
7KR6
 
 | |
4PQ9
 
 | |
5SV8
 
 | | Crystal Structure of the catalytic nucleophile and surface cysteine mutant of VvEG16 in complex with a xyloglucan oligosaccharide | | Descriptor: | alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, probable xyloglucan endotransglucosylase/hydrolase protein 19 | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
3WVJ
 
 | | The crystal structure of native glycosidic hydrolase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | Authors: | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
1U0A
 
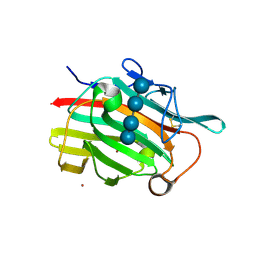 | | Crystal structure of the engineered beta-1,3-1,4-endoglucanase H(A16-M) in complex with beta-glucan tetrasaccharide | | Descriptor: | Beta-glucanase, CALCIUM ION, ZINC ION, ... | | Authors: | Gaiser, O.J, Piotukh, K, Ponnuswamy, M.N, Planas, A, Borriss, R, Heinemann, U. | | Deposit date: | 2004-07-13 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis for the Substrate Specificity of a Bacillus 1,3-1,4-beta-Glucanase
J.Mol.Biol., 357, 2006
|
|
4XXP
 
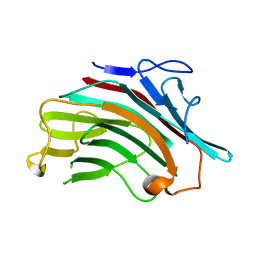 | |
1CPN
 
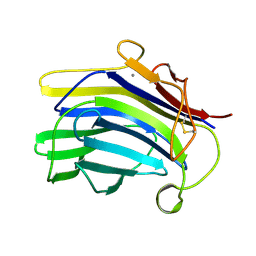 | |
4XDQ
 
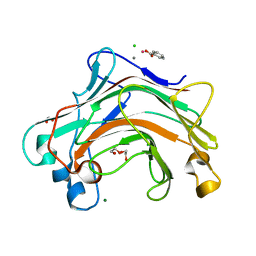 | |
4W65
 
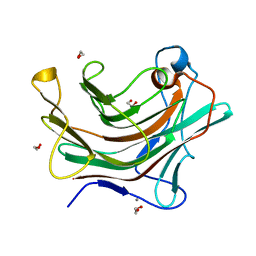 | |
8EP4
 
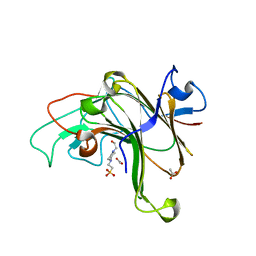 | | Structure of Bacple_01703 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
1CPM
 
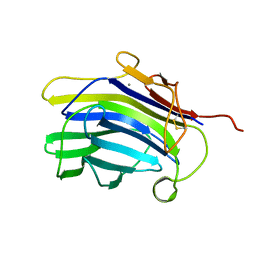 | |
4WZF
 
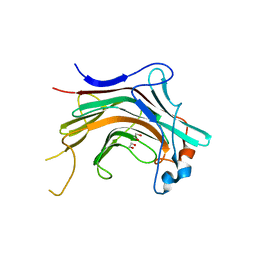 | | Crystal structural basis for Rv0315, an immunostimulatory antigen and pseudo beta-1, 3-glucanase of Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanase, CALCIUM ION | | Authors: | Dong, W.Y, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structural basis for Rv0315, an immunostimulatory antigen and inactive beta-1,3-glucanase of Mycobacterium tuberculosis.
Sci Rep, 5, 2015
|
|
3O5S
 
 | | Crystal Structure of the endo-beta-1,3-1,4 glucanase from Bacillus subtilis (strain 168) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Santos, C.R, Tonoli, C.C.C, Souza, A.R, Furtado, G.P, Ribeiro, L.F, Ward, R.J, Murakami, M.T. | | Deposit date: | 2010-07-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a Beta-1,3 1,4-glucanase from Bacillus subtilis 168
PROCESS BIOCHEM, 46, 2011
|
|
8EW1
 
 | | Structure of Bacple_01703-E145L | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
1MVE
 
 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
3RQ0
 
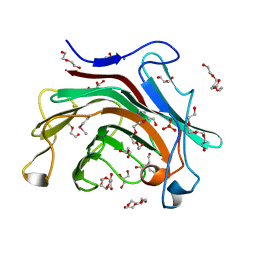 | | The crystal structure of a glycosyl hydrolases (GH) family protein 16 from Mycobacterium smegmatis str. MC2 155 | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of a glycosyl hydrolases (GH) family protein 16 from Mycobacterium smegmatis str. MC2 155
To be Published
|
|
1DYP
 
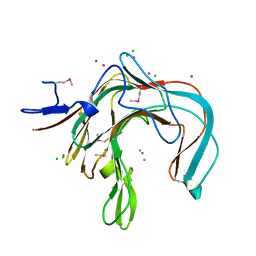 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
6IBU
 
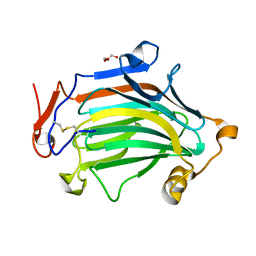 | | Apo Crh5 transglycosylase | | Descriptor: | GLYCEROL, Probable glycosidase crf1 | | Authors: | Bartual, S.G, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
6IBW
 
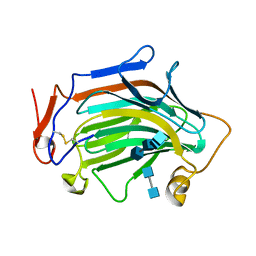 | | Crh5 transglycosylase in complex with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable glycosidase crf1 | | Authors: | Fang, W, Bartual, S.G, van Aalten, D.M.F. | | Deposit date: | 2018-12-01 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
