8SOT
 
 | |
5GPH
 
 | |
5EZ1
 
 | |
6BHF
 
 | |
8HM4
 
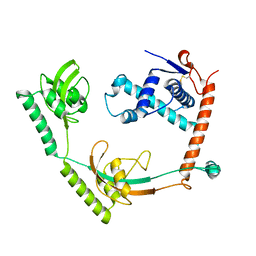 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
6DUN
 
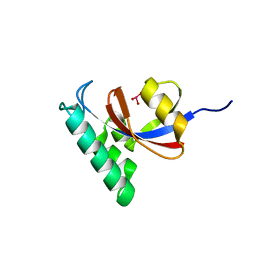 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, TRIHYDROXYARSENITE(III) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells.
Nat Commun, 9, 2018
|
|
1J6Y
 
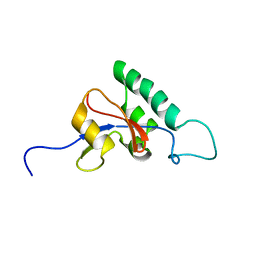 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
5TVL
 
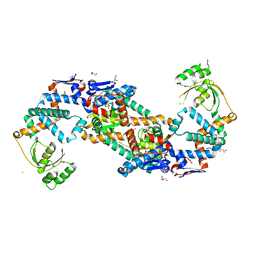 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
6VJ4
 
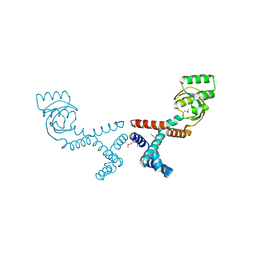 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6GMP
 
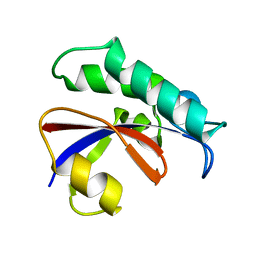 | | CRYSTAL STRUCTURE OF THE PPIASE DOMAIN OF TBPAR42 | | Descriptor: | PARVULIN 42 | | Authors: | Hoenig, D, Rute, A, Hofmann, E, Bayer, P, Gasper, R. | | Deposit date: | 2018-05-28 | | Release date: | 2019-03-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of the 42 kDa Parvulin ofTrypanosoma brucei.
Biomolecules, 9, 2019
|
|
1NMW
 
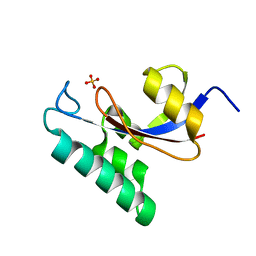 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
2JZV
 
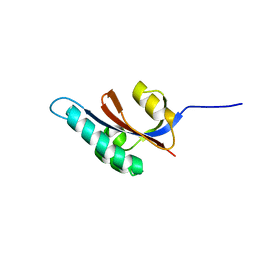 | | Solution structure of S. aureus PrsA-PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Seppala, R, Tossavainen, H, Heikkinen, S, Koskela, H, Kontinen, V, Permi, P. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA - Implications for the catalytic mechanism of parvulins.
Bmc Struct.Biol., 9, 2009
|
|
2LJ4
 
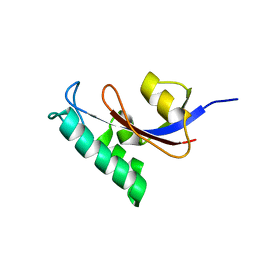 | | Solution structure of the TbPIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase/rotamase, putative | | Authors: | Sun, L, Lin, D, Zhao, Y. | | Deposit date: | 2011-09-06 | | Release date: | 2012-08-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structural analysis of the single-domain parvulin TbPin1.
Plos One, 7, 2012
|
|
4TNS
 
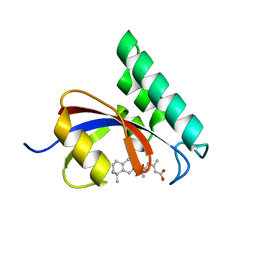 | |
4TYO
 
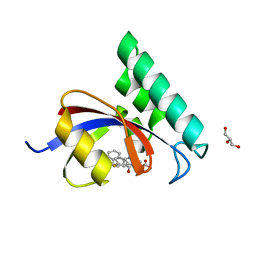 | | PPIase in complex with a non-phosphate small molecule inhibitor. | | Descriptor: | 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-D-alanine, GLYCEROL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2N87
 
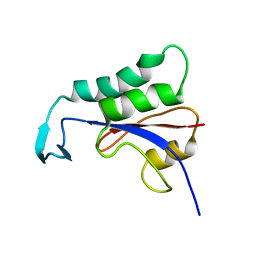 | |
2PV2
 
 | |
2PV1
 
 | |
2RUC
 
 | |
2RUQ
 
 | | solution structure of human Pin1 PPIase mutant C113A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
2RUD
 
 | |
2RUR
 
 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
3GPK
 
 | |
3I6C
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | 3-fluoro-N-(naphthalen-2-ylcarbonyl)-D-phenylalanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3IKD
 
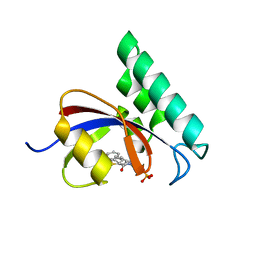 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
