8Y6O
 
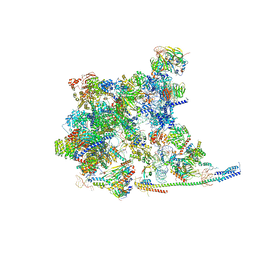 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
8XPN
 
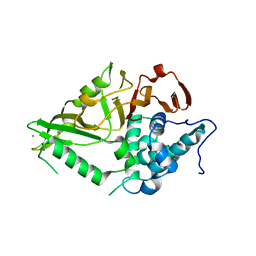 | | The Crystal Structure of USP8 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
8WG5
 
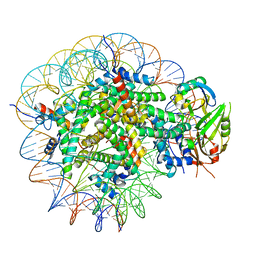 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome
To Be Published
|
|
8OYP
 
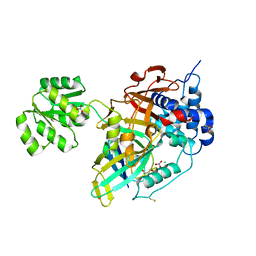 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
8ITP
 
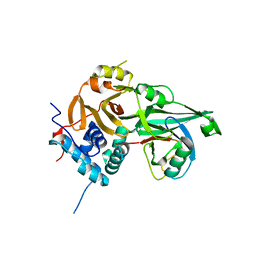 | |
8ITN
 
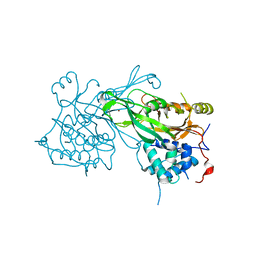 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design
Commun Biol, 6, 2023
|
|
8HJE
 
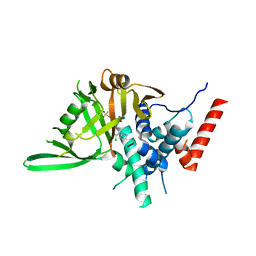 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
8D4Z
 
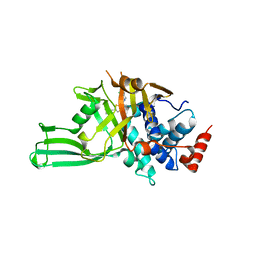 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | Descriptor: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bell, J.A. | | Deposit date: | 2022-06-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
8D1T
 
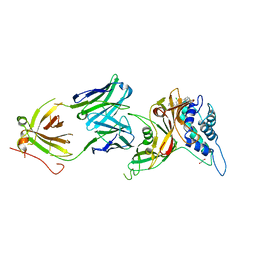 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
8D0A
 
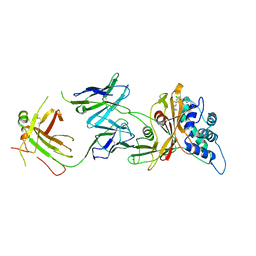 | | Crystal structure of human USP30 in complex with a covalent inhibitor 829 and a Fab | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, mouse anti-huUSP30 Fab heavy chain, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | TBD
To Be Published
|
|
8BS9
 
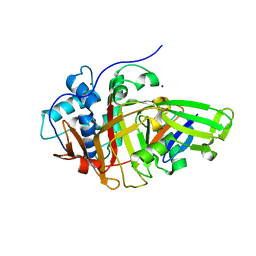 | | Structure of USP36 in complex with Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 36, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8BS3
 
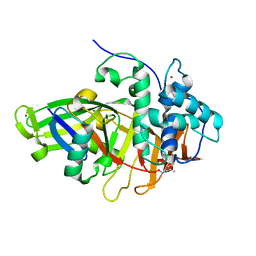 | | Structure of USP36 in complex with Fubi-PA | | Descriptor: | 40S ribosomal protein S30, Ubiquitin carboxyl-terminal hydrolase 36, ZINC ION, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8A9K
 
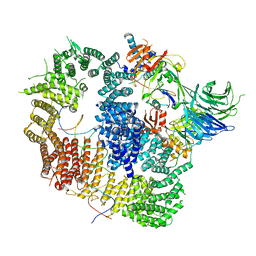 | | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, DNA (61-MER), Fanconi anemia group D2 protein, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
8A9J
 
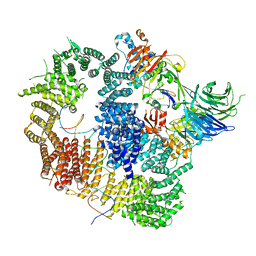 | |
7ZH4
 
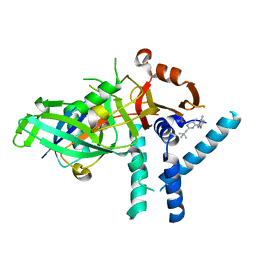 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7ZH3
 
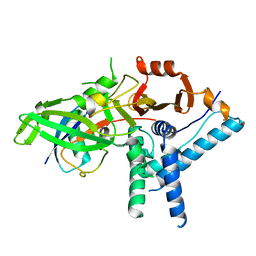 | |
7YXY
 
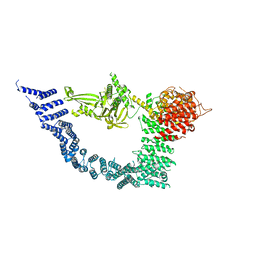 | |
7YXX
 
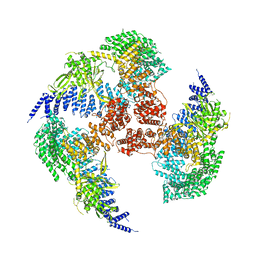 | |
7XHK
 
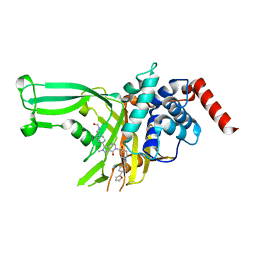 | | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[[4-[6-[[1-[[2-chloranyl-4-(furan-2-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-3-yl]phenyl]methyl]methanamide | | Authors: | Sun, H.B, Wen, X.A. | | Deposit date: | 2022-04-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46
To Be Published
|
|
7XHH
 
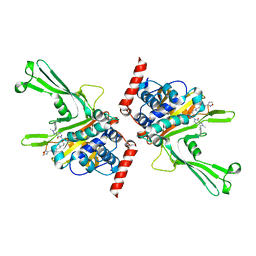 | | High-resolution X-ray cocrystal structure of USP7 in complex with X4 | | Descriptor: | 3-[4-(aminomethyl)phenyl]-6-[[1-[[2-chloranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-pyrazolo[4,3-d]pyrimidin-7-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Sun, H.B, Wen, X.A. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution X-ray cocrystal structure of USP7 in complex with X4
To Be Published
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3R
 
 | | USP34 catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3M
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3K
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3J
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
