1JE8
 
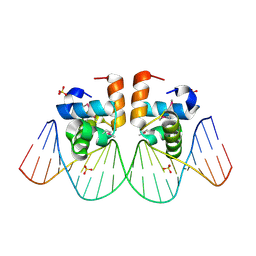 | | Two-Component response regulator NarL/DNA Complex: DNA Bending Found in a High Affinity Site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/Nitrite Response Regulator Protein NARL, SULFATE ION | | Authors: | Maris, A.E, Sawaya, M.R, Kaczor-Grzeskowiak, M, Jarvis, M.R, Bearson, S.M.D, Kopka, M.L, Schroder, I, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2001-06-15 | | Release date: | 2002-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dimerization allows DNA target site recognition by the NarL response regulator.
Nat.Struct.Biol., 9, 2002
|
|
1P4W
 
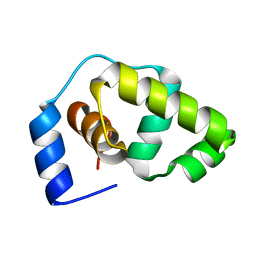 | | Solution structure of the DNA-binding domain of the Erwinia amylovora RcsB protein | | Descriptor: | rcsB | | Authors: | Pristovsek, P, Sengupta, K, Loehr, F, Schaefer, B, Wehland von Trebra, M, Rueterjans, H, Bernhard, F. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box.
J.Biol.Chem., 278, 2003
|
|
1FSE
 
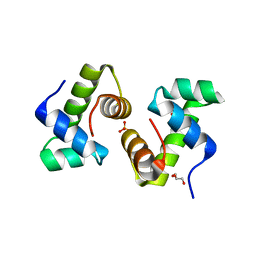 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
7VE5
 
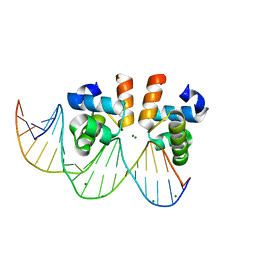 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
2JPC
 
 | |
2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
2KRF
 
 | | NMR solution structure of the DNA binding domain of Competence protein A | | Descriptor: | Transcriptional regulatory protein comA | | Authors: | Hobbs, C.A, Bobay, B.G, Thompson, R.J, Perego, M, Cavanagh, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and DNA-binding model of the DNA-binding domain of competence protein A.
J.Mol.Biol., 398, 2010
|
|
3C57
 
 | |
3CLO
 
 | |
7VIM
 
 | |
7X1K
 
 | |
6JQS
 
 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
1X3U
 
 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
1ZLK
 
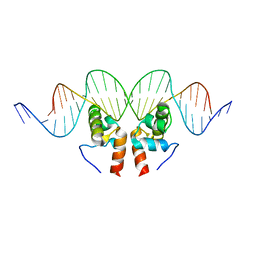 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR C-terminal Domain-DNA Complex | | Descriptor: | 5'-D(*CP*GP*TP*GP*GP*CP*CP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*AP*CP*TP*TP*TP*AP*GP*TP*CP*CP*CP*CP*AP*AP*AP*GP*CP*GP*CP*GP*GP*GP*CP*CP*AP*T)-3', 5'-D(*GP*GP*CP*CP*CP*GP*CP*GP*CP*TP*TP*TP*GP*GP*GP*GP*AP*CP*TP*AP*AP*AP*GP*TP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*GP*GP*CP*CP*AP*CP*GP*AP*T)-3', Dormancy Survival Regulator | | Authors: | Wisedchaisri, G, Wu, M, Rice, A.E, Roberts, D.M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency.
J.Mol.Biol., 354, 2005
|
|
1ZG5
 
 | | NarL complexed to narG-89 promoter palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*CP*TP*AP*TP*AP*GP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, SULFATE ION | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1ZLJ
 
 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR C-terminal Domain | | Descriptor: | Dormancy Survival Regulator | | Authors: | Wisedchaisri, G, Wu, M, Rice, A.E, Roberts, D.M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency.
J.Mol.Biol., 354, 2005
|
|
1ZG1
 
 | | NarL complexed to nirB promoter non-palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*GP*GP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*CP*CP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, ... | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
7DWM
 
 | | Crystal structure of the phage VqmA-DPO complex | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | Authors: | Gu, Y, Yang, W.S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
4LGW
 
 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | Descriptor: | GLYCEROL, Regulatory protein SdiA | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SZT
 
 | | Quorum Sensing Control Repressor, QscR, Bound to N-3-oxo-dodecanoyl-L-Homoserine Lactone | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Quorum-sensing control repressor, SODIUM ION | | Authors: | Churchill, M.E.A, Lintz, M.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ULQ
 
 | |
2Q0O
 
 | | Crystal structure of an anti-activation complex in bacterial quorum sensing | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Probable transcriptional activator protein traR, Probable transcriptional repressor traM | | Authors: | Chen, G, Jeffrey, P.D, Fuqua, C, Shi, Y, Chen, L. | | Deposit date: | 2007-05-22 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antiactivation in bacterial quorum sensing.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1L3L
 
 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
3KLN
 
 | | Vibrio cholerae VpsT | | Descriptor: | Transcriptional regulator, LuxR family | | Authors: | Krasteva, P.V, Navarro, V.A.S, Sondermann, H. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP.
Science, 327, 2010
|
|
1H0M
 
 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
